Dissecting the evolutionary role of the Hox gene proboscipedia in Drosophila mouthpart diversification by full locus replacement
- PMID: 34757777
- PMCID: PMC8580299
- DOI: 10.1126/sciadv.abk1003
Dissecting the evolutionary role of the Hox gene proboscipedia in Drosophila mouthpart diversification by full locus replacement
Abstract
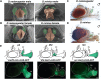
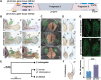
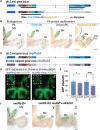
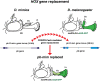
Hox genes determine positional codes along the head-to-tail axis. Here, we replaced the entire Drosophila melanogaster proboscipedia (pb) Hox locus, which controls the development of the proboscis and maxillary palps, with that from Drosophila mimica, a related species with highly modified mouthparts. The D. mimica replacement rescues most aspects of adult proboscis morphology; however, the shape and orientation of maxillary palps were modified, resembling D. mimica and closely related species. Expressing the D. mimica Pb protein in the D. melanogaster pattern fully rescued D. melanogaster morphology. However, the expression pattern directed by D. mimica pb cis-regulatory sequences differed from that of D. melanogaster pb in cells that produce altered maxillary structures, indicating that pb regulatory sequences can evolve in related species to alter morphology.
Figures
Similar articles
-
Genetic characterization of the role of the two HOX proteins, Proboscipedia and Sex Combs Reduced, in determination of adult antennal, tarsal, maxillary palp and proboscis identities in Drosophila melanogaster.Development. 1997 Dec;124(24):5049-62. doi: 10.1242/dev.124.24.5049. Development. 1997. PMID: 9362475
-
The Drosophila proboscis is specified by two Hox genes, proboscipedia and Sex combs reduced, via repression of leg and antennal appendage genes.Development. 2001 Jul;128(14):2803-14. doi: 10.1242/dev.128.14.2803. Development. 2001. PMID: 11526085
-
RNAi analysis of Deformed, proboscipedia and Sex combs reduced in the milkweed bug Oncopeltus fasciatus: novel roles for Hox genes in the hemipteran head.Development. 2000 Sep;127(17):3683-94. doi: 10.1242/dev.127.17.3683. Development. 2000. PMID: 10934013
-
Understanding the genetic basis of morphological evolution: the role of homeotic genes in the diversification of the arthropod bauplan.Int J Dev Biol. 1998;42(3):453-61. Int J Dev Biol. 1998. PMID: 9654031 Review.
-
Lessons on gene regulation learnt from the Drosophila melanogaster bithorax complex.Int J Dev Biol. 2020;64(1-2-3):151-158. doi: 10.1387/ijdb.190167rm. Int J Dev Biol. 2020. PMID: 32659003 Review.
Cited by
-
HOXA10 and HOXA11 in Human Endometrial Benign Disorders: Unraveling Molecular Pathways and Their Impact on Reproduction.Biomolecules. 2025 Apr 10;15(4):563. doi: 10.3390/biom15040563. Biomolecules. 2025. PMID: 40305321 Free PMC article. Review.
-
Active genetics comes alive: Exploring the broad applications of CRISPR-based selfish genetic elements (or gene-drives): Exploring the broad applications of CRISPR-based selfish genetic elements (or gene-drives).Bioessays. 2022 Aug;44(8):e2100279. doi: 10.1002/bies.202100279. Epub 2022 Jun 9. Bioessays. 2022. PMID: 35686327 Free PMC article. Review.
-
Hox gene-specific cellular targeting using split intein Trojan exons.Proc Natl Acad Sci U S A. 2024 Apr 23;121(17):e2317083121. doi: 10.1073/pnas.2317083121. Epub 2024 Apr 11. Proc Natl Acad Sci U S A. 2024. PMID: 38602904 Free PMC article.
References
-
- Carroll S. B., Homeotic genes and the evolution of arthropods and chordates. Nature 376, 479–485 (1995). - PubMed
-
- Di Gregorio A., Spagnuolo A., Ristoratore F., Pischetola M., Aniello F., Branno M., Cariello L., Lauro R. D., Cloning of ascidian homeobox genes provides evidence for a primordial chordate cluster. Gene 156, 253–257 (1995). - PubMed
-
- Spagnuolo A., Ristoratore F., di Gregorio A., Aniello F., Branno M., di Lauro R., Unusual number and genomic organization of Hox genes in the tunicate Ciona intestinalis. Gene 309, 71–79 (2003). - PubMed
-
- Wada H., Garcia-Fernàndez J., Holland P. W. H., Colinear and segmental expression of amphioxus Hox genes. Dev. Biol. 213, 131–141 (1999). - PubMed
-
- Meyer A., Málaga-Trillo E., Vertebrate genomics: More fishy tales about Hox genes. Curr. Biol. 9, R210–R213 (1999). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

