Subcellular Localization Defects Characterize Ribose-Binding Mutant Proteins with New Ligand Properties in Escherichia coli
- PMID: 34757821
- PMCID: PMC8788693
- DOI: 10.1128/AEM.02117-21
Subcellular Localization Defects Characterize Ribose-Binding Mutant Proteins with New Ligand Properties in Escherichia coli
Abstract
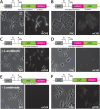
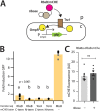
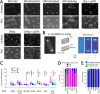
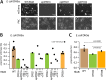
Periplasmic binding proteins have been previously proclaimed as a general scaffold to design sensor proteins with new recognition specificities for nonnatural compounds. Such proteins can be integrated in bacterial bioreporter chassis with hybrid chemoreceptors to produce a concentration-dependent signal after ligand binding to the sensor cell. However, computationally designed new ligand-binding properties ignore the more general properties of periplasmic binding proteins, such as their periplasmic translocation, dynamic transition of open and closed forms, and interactions with membrane receptors. In order to better understand the roles of such general properties in periplasmic signaling behavior, we studied the subcellular localization of ribose-binding protein (RbsB) in Escherichia coli in comparison to a recently evolved set of mutants designed to bind 1,3-cyclohexanediol. As proxies for localization, we calibrated and deployed C-terminal end mCherry fluorescent protein fusions. Whereas RbsB-mCherry coherently localized to the periplasmic space and accumulated in (periplasmic) polar regions depending on chemoreceptor availability, mutant RbsB-mCherry expression resulted in high fluorescence cell-to-cell variability. This resulted in higher proportions of cells devoid of clear polar foci and of cells with multiple fluorescent foci elsewhere, suggesting poorer translocation, periplasmic autoaggregation, and mislocalization. Analysis of RbsB mutants and mutant libraries at different stages of directed evolution suggested overall improvement to more RbsB-wild-type-like characteristics, which was corroborated by structure predictions. Our results show that defects in periplasmic localization of mutant RbsB proteins partly explain their poor sensing performance. Future efforts should be directed to predicting or selecting secondary mutations outside computationally designed binding pockets, taking folding, translocation, and receptor interactions into account. IMPORTANCE Biosensor engineering relies on transcription factors or signaling proteins to provide the actual sensory functions for the target chemicals. Since for many compounds there are no natural sensory proteins, there is a general interest in methods that could unlock routes to obtaining new ligand-binding properties. Bacterial periplasmic binding proteins (PBPs) form an interesting family of proteins to explore for this purpose, because there is a large natural variety suggesting evolutionary trajectories to bind new ligands. PBPs are conserved and amenable to accurate computational binding pocket predictions. However, studying ribose-binding protein in Escherichia coli, we discovered that designed variants have defects in their proper localization in the cell, which can impair appropriate sensor signaling. This indicates that functional sensing capacity of PBPs cannot be obtained solely through computational design of the ligand-binding pocket but must take other properties of the protein into account, which are currently very difficult to predict.
Keywords: biosensing; fluorescent protein fusion; protein localization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

