Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2
- PMID: 34758478
- PMCID: PMC8732273
- DOI: 10.1038/s41586-021-04186-8
Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2
Abstract
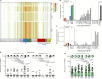
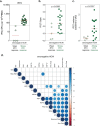
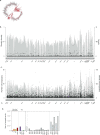
Individuals with potential exposure to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) do not necessarily develop PCR or antibody positivity, suggesting that some individuals may clear subclinical infection before seroconversion. T cells can contribute to the rapid clearance of SARS-CoV-2 and other coronavirus infections1-3. Here we hypothesize that pre-existing memory T cell responses, with cross-protective potential against SARS-CoV-2 (refs. 4-11), would expand in vivo to support rapid viral control, aborting infection. We measured SARS-CoV-2-reactive T cells, including those against the early transcribed replication-transcription complex (RTC)12,13, in intensively monitored healthcare workers (HCWs) who tested repeatedly negative according to PCR, antibody binding and neutralization assays (seronegative HCWs (SN-HCWs)). SN-HCWs had stronger, more multispecific memory T cells compared with a cohort of unexposed individuals from before the pandemic (prepandemic cohort), and these cells were more frequently directed against the RTC than the structural-protein-dominated responses observed after detectable infection (matched concurrent cohort). SN-HCWs with the strongest RTC-specific T cells had an increase in IFI27, a robust early innate signature of SARS-CoV-2 (ref. 14), suggesting abortive infection. RNA polymerase within RTC was the largest region of high sequence conservation across human seasonal coronaviruses (HCoV) and SARS-CoV-2 clades. RNA polymerase was preferentially targeted (among the regions tested) by T cells from prepandemic cohorts and SN-HCWs. RTC-epitope-specific T cells that cross-recognized HCoV variants were identified in SN-HCWs. Enriched pre-existing RNA-polymerase-specific T cells expanded in vivo to preferentially accumulate in the memory response after putative abortive compared to overt SARS-CoV-2 infection. Our data highlight RTC-specific T cells as targets for vaccines against endemic and emerging Coronaviridae.
© 2021. The Author(s).
Conflict of interest statement
A.B. is a cofounder of Lion TCR, a biotechnology company that develops T cell receptors for the treatment of virus-related diseases and cancers. R.J.B. and D.M.A. are members of the Global T-cell Expert Consortium and have consulted for Oxford Immunotec outside the submitted work. The other authors declare no competing interests.
Figures









Comment in
-
How do people resist COVID infections? Hospital workers offer a hint.Nature. 2021 Nov;599(7886):543. doi: 10.1038/d41586-021-03110-4. Nature. 2021. PMID: 34773110 No abstract available.
-
Cross-reactive memory T cells abort SARS-CoV-2 infection.Nat Rev Immunol. 2022 Jan;22(1):5. doi: 10.1038/s41577-021-00667-z. Nat Rev Immunol. 2022. PMID: 34873280 Free PMC article. No abstract available.
-
In the shadow of antibodies: how T cells defend against COVID-19.Ann Rheum Dis. 2022 Jun;81(6):757-759. doi: 10.1136/annrheumdis-2021-221952. Epub 2022 Apr 7. Ann Rheum Dis. 2022. PMID: 35393270 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- MC_PC_20031/MRC_/Medical Research Council/United Kingdom
- MR/S019553/1/MRC_/Medical Research Council/United Kingdom
- 26603/CRUK_/Cancer Research UK/United Kingdom
- MR/W020610/1/MRC_/Medical Research Council/United Kingdom
- FS/CRTF/21/24128/BHF_/British Heart Foundation/United Kingdom
- 207511/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- MC_PC_20060/MRC_/Medical Research Council/United Kingdom
- MR/V036939/1/MRC_/Medical Research Council/United Kingdom
- MR/R02622X/1/MRC_/Medical Research Council/United Kingdom
- MR/R008698/1/MRC_/Medical Research Council/United Kingdom
- MRF-044-0001-RG-SWADL/MRF_/MRF_/United Kingdom
- 214191/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- FS/18/21/33447/BHF_/British Heart Foundation/United Kingdom
- MR/V027883/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

