Optimization of lipase production using fungal isolates from oily residues
- PMID: 34758800
- PMCID: PMC8582195
- DOI: 10.1186/s12896-021-00724-4
Optimization of lipase production using fungal isolates from oily residues
Abstract
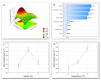
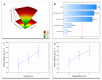
Lipases are triacylglycerol hydrolases that catalyze hydrolysis, esterification, interesterification, and transesterification reactions. These enzymes are targets of several industrial and biotech applications, such as catalysts, detergent production, food, biofuels, wastewater treatment, and others. Microbial enzymes are preferable for large scale production due to ease of production and extraction. Several studies have reported that lipases from filamentous fungi are predominantly extracellular and highly active. However, there are many factors that interfere with enzyme production (pH, temperature, medium composition, agitation, aeration, inducer type, and concentration, etc.), making control difficult and burdening the process. This work aimed to optimize the lipase production of four fungal isolates from oily residues (Penicillium sp., Aspergillus niger, Aspergillus sp., and Aspergillus sp.). The lipase-producing fungi isolates were morphologically characterized by optical and scanning electron microscopy. The optimal lipase production time curve was previously determined, and the response variable used was the amount of total protein in the medium after cultivation by submerged fermentation. A complete factorial design 32 was performed, evaluating the temperatures (28 °C, 32 °C, and 36 °C) and soybean oil inducer concentration (2%, 6%, and 10%). Each lipase-producing isolate reacted differently to the conditions tested, the Aspergillus sp. F18 reached maximum lipase production, compared to others, under conditions of 32 °C and 2% of oil with a yield of 11,007 (µg mL-1). Penicillium sp. F04 achieved better results at 36 °C and 6% oil, although for Aspergillus niger F16 was at 36 °C and 10% oil and Aspergillus sp. F21 at 32 °C and 2% oil. These results show that microorganisms isolated from oily residues derived from environmental sanitation can be a promising alternative for the large-scale production of lipases.
Keywords: Factorial design; Fungal lipases; Oily waste; Optimization.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Gonçalves CCS, Marsaioli AJ. Facts and trends of biocatalysis. Quim Nova. 2013;36:1587–1590. doi: 10.1590/S0100-40422013001000016. - DOI
-
- Jegannathan KR, Nielsen PH. Environmental assessment of enzyme use in industrial production—a literature review. J Clean Prod. 2013 doi: 10.1016/j.jclepro.2012.11.005. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

