Self-guarding of MORC3 enables virulence factor-triggered immunity
- PMID: 34759314
- PMCID: PMC9045311
- DOI: 10.1038/s41586-021-04054-5
Self-guarding of MORC3 enables virulence factor-triggered immunity
Abstract
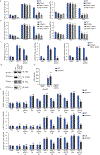
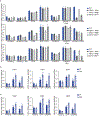
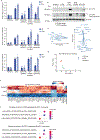
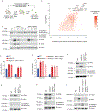
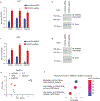
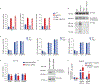
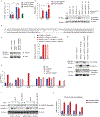
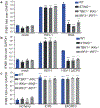
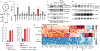
Pathogens use virulence factors to inhibit the immune system1. The guard hypothesis2,3 postulates that hosts monitor (or 'guard') critical innate immune pathways such that their disruption by virulence factors provokes a secondary immune response1. Here we describe a 'self-guarded' immune pathway in human monocytes, in which guarding and guarded functions are combined in one protein. We find that this pathway is triggered by ICP0, a key virulence factor of herpes simplex virus type 1, resulting in robust induction of anti-viral type I interferon (IFN). Notably, induction of IFN by ICP0 is independent of canonical immune pathways and the IRF3 and IRF7 transcription factors. A CRISPR screen identified the ICP0 target MORC34 as an essential negative regulator of IFN. Loss of MORC3 recapitulates the IRF3- and IRF7-independent IFN response induced by ICP0. Mechanistically, ICP0 degrades MORC3, which leads to de-repression of a MORC3-regulated DNA element (MRE) adjacent to the IFNB1 locus. The MRE is required in cis for IFNB1 induction by the MORC3 pathway, but is not required for canonical IFN-inducing pathways. As well as repressing the MRE to regulate IFNB1, MORC3 is also a direct restriction factor of HSV-15. Our results thus suggest a model in which the primary anti-viral function of MORC3 is self-guarded by its secondary IFN-repressing function-thus, a virus that degrades MORC3 to avoid its primary anti-viral function will unleash the secondary anti-viral IFN response.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
REV consults for Ventus Therapeutics and Tempest Therapeutics. A provisional patent application covering these findings has been submitted.
Figures


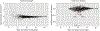


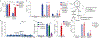

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

