Detection of MSH 2 Gene Methylation in Extramammary Paget's Disease by Methylation-Sensitive High-Resolution Melting Analysis
- PMID: 34759969
- PMCID: PMC8575627
- DOI: 10.1155/2021/5514426
Detection of MSH 2 Gene Methylation in Extramammary Paget's Disease by Methylation-Sensitive High-Resolution Melting Analysis
Abstract
Background: Extramammary Paget's disease (EMPD) is a rare skin tumor. Hypermethylation in the MSH2 promoter resulting in the downregulation of its protein expression shows a high detection rate in EMPD tumor tissue, which indicates that the methylation of MSH2 may play an important role in the pathogenesis of EMPD.
Objective: This study aims to establish a rapid analysis strategy based on the methylation-sensitive high-resolution melting curve (MS-HRM) to detect the methylation level of the MSH2 promoter.
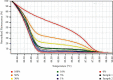
Methods: With the use of universal methylated human DNA products, we established the MS-HRM standard curve to quantitatively detect the methylation level of the MSH2 promoter. Then, all 57 EMPD tumor DNA samples were analyzed. Pyrosequencing assay was also carried out to test the accuracy and efficacy of MS-HRM. Besides, a total of 54 human normal and other cancerous tissues were included in this study to test the reliability and versatility of the MS-HRM standard curve.
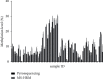
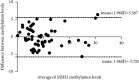
Results: In this study, by using the established MS-HRM, we found that 96.5% (55/57) EMPD tumor samples had varying methylation levels in the MSH2 promoter ranging from 0% to 30%. Then, the methylation data were compared to the results obtained from pyrosequencing, which showed a high correlation between these two techniques by Pearson's correlation (r = 0.9425) and Bland-Altman plots (mean difference = -0.1069) indicating that the methylation levels analyzed by MS-HRM were consistent with DNA pyrosequencing. Furthermore, in 23 normal and 31 other cancerous tissue samples, there were two colorectal cancer (CRC) tissues that tested MSH2 methylation positive (1% and 5%) which confirmed that our established MS-HRM can be widely applied to various types of samples.
Conclusion: MS-HRM standard curve can be used for the detection of the methylation level of MSH2 in EMPD tumor samples and other cancerous tissues potentially, which presents a promising candidate as a quantitative assay to analyze MSH2 promoter methylation in routine pathological procedure.
Copyright © 2021 Liu Dong et al.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Whole genome methylation sequencing reveals epigenetic landscape and abnormal expression of FABP5 in extramammary Paget's disease.Skin Res Technol. 2023 Oct;29(10):e13497. doi: 10.1111/srt.13497. Skin Res Technol. 2023. PMID: 37881057 Free PMC article.
-
Methylation and expression analysis of mismatch repair genes in extramammary Paget's disease.J Eur Acad Dermatol Venereol. 2019 May;33(5):874-879. doi: 10.1111/jdv.15404. Epub 2019 Feb 19. J Eur Acad Dermatol Venereol. 2019. PMID: 30784122
-
Rapid determination of AKAP12 promoter methylation levels in peripheral blood using methylation-sensitive high resolution melting (MS-HRM) analysis: application in colorectal cancer.Clin Chim Acta. 2010 Jul 4;411(13-14):940-6. doi: 10.1016/j.cca.2010.03.003. Epub 2010 Mar 11. Clin Chim Acta. 2010. PMID: 20227403
-
Definition, Association with Malignancy, Biologic Behavior, and Treatment of Ectopic Extramammary Paget's Disease: A Review of the Literature.J Clin Aesthet Dermatol. 2019 Aug;12(8):40-44. Epub 2019 Aug 1. J Clin Aesthet Dermatol. 2019. PMID: 31531170 Free PMC article. Review.
-
Extramammary Paget's disease: what do we know and how do we treat?Can J Urol. 2019 Dec;26(6):10012-10021. Can J Urol. 2019. PMID: 31860417 Review.
Cited by
-
Whole genome methylation sequencing reveals epigenetic landscape and abnormal expression of FABP5 in extramammary Paget's disease.Skin Res Technol. 2023 Oct;29(10):e13497. doi: 10.1111/srt.13497. Skin Res Technol. 2023. PMID: 37881057 Free PMC article.
References
LinkOut - more resources
Full Text Sources

