Recent developments in chemical conjugation strategies targeting native amino acids in proteins and their applications in antibody-drug conjugates
- PMID: 34760149
- PMCID: PMC8549674
- DOI: 10.1039/d1sc02973h
Recent developments in chemical conjugation strategies targeting native amino acids in proteins and their applications in antibody-drug conjugates
Abstract
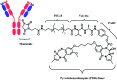
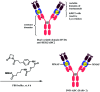
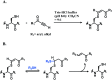
Many fields in chemical biology and synthetic biology require effective bioconjugation methods to achieve their desired functions and activities. Among such biomolecule conjugates, antibody-drug conjugates (ADCs) need a linker that provides a stable linkage between cytotoxic drugs and antibodies, whilst conjugating in a biologically benign, fast and selective fashion. This review focuses on how the development of novel organic synthesis can solve the problems of traditional linker technology. The review shall introduce and analyse the current developments in the modification of native amino acids on peptides or proteins and their applicability to ADC linker. Thereafter, the review shall discuss in detail each endogenous amino acid's intrinsic reactivity and selectivity aspects, and address the research effort to construct an ADC using each conjugation method.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

























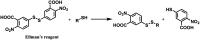

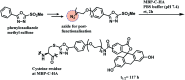













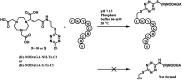





















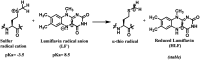





References
-
- Bross P. Beitz J. Chen G. Chen X. Duffy E. Keiffer L. Roy S. Sridhara R. Rahman A. Williams G. Pazdur R. Clin. Cancer Res. 2001;7:1490–1496. - PubMed
-
- de Claro R. A. McGinn K. Kwitkowski V. Bullock J. Khandelwal A. Habtemariam B. Ouyang Y. Saber H. Lee K. Koti K. Rothmann M. Shapiro M. Borrego F. Clouse K. Chen X. H. Brown J. Akinsanya L. Kane R. Kaminskas E. Farrell A. Pazdur R. Clin. Cancer Res. 2012;18:5845–5849. doi: 10.1158/1078-0432.CCR-12-1803. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

