An Interactive Pipeline for Quantitative Histopathological Analysis of Spatially Defined Drug Effects in Tumors
- PMID: 34760331
- PMCID: PMC8529341
- DOI: 10.4103/jpi.jpi_17_21
An Interactive Pipeline for Quantitative Histopathological Analysis of Spatially Defined Drug Effects in Tumors
Abstract
Background: Tumor heterogeneity is increasingly being recognized as a major source of variability in the histopathological assessment of drug responses. Quantitative analysis of immunohistochemistry (IHC) and immunofluorescence (IF) images using biomarkers that capture spatialpatterns of distinct tumor biology and drug concentration in tumors is of high interest to the field.
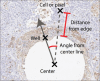
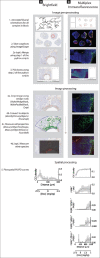
Methods: We have developed an image analysis pipeline to measure drug response using IF and IHC images along spatial gradients of local drug release from a tumor-implantable drug delivery microdevice. The pipeline utilizes a series of user-interactive python scripts and CellProfiler pipelines with custom modules to perform image and spatial analysis of regions of interest within whole-slide images.
Results: Worked examples demonstrate that intratumor measurements such as apoptosis, cell proliferation, and immune cell population density can be quantitated in a spatially and drug concentration-dependent manner, establishing in vivo profiles of pharmacodynamics and pharmacokinetics in tumors.
Conclusions: Spatial image analysis of tumor response along gradients of local drug release is achievable in high throughput. The major advantage of this approach is the use of spatially aware annotation tools to correlate drug gradients with drug effects in tumors in vivo.
Keywords: Controlled release; digital pathology; image annotation; quantitative pathology; spatial analysis; tumor microenvironment.
Copyright: © 2021 Journal of Pathology Informatics.
Conflict of interest statement
There are no conflicts of interest.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

