Evolutionary history of the vertebrate Piwi gene family
- PMID: 34760405
- PMCID: PMC8574217
- DOI: 10.7717/peerj.12451
Evolutionary history of the vertebrate Piwi gene family
Abstract
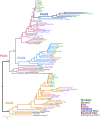
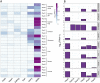
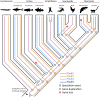
PIWIs are regulatory proteins that belong to the Argonaute family. Piwis are primarily expressed in gonads and protect the germline against the mobilization and propagation of transposable elements (TEs) through transcriptional gene silencing. Vertebrate genomes encode up to four Piwi genes: Piwil1, Piwil2, Piwil3 and Piwil4, but their duplication history is unresolved. We leveraged phylogenetics, synteny and expression analyses to address this void. Our phylogenetic analysis suggests Piwil1 and Piwil2 were retained in all vertebrate members. Piwil4 was the result of Piwil1 duplication in the ancestor of gnathostomes, but was independently lost in ray-finned fishes and birds. Further, Piwil3 was derived from a tandem Piwil1 duplication in the common ancestor of marsupial and placental mammals, but was secondarily lost in Atlantogenata (Xenarthra and Afrotheria) and some rodents. The evolutionary rate of Piwil3 is considerably faster than any Piwi among all lineages, but an explanation is lacking. Our expression analyses suggest Piwi expression has mostly been constrained to gonads throughout vertebrate evolution. Vertebrate evolution is marked by two early rounds of whole genome duplication and many multigene families are linked to these events. However, our analyses suggest Piwi expansion was independent of whole genome duplications.
Keywords: Argonaute gene family; Gene duplication; Phylogenetics; RNAi; Selection; Synteny; Transcriptomics.
©2021 Gutierrez et al.
Conflict of interest statement
David Ray is an Academic Editor for PeerJ.
Figures


References
LinkOut - more resources
Full Text Sources

