Genome analysis suggests the bacterial family Acetobacteraceae is a source of undiscovered specialized metabolites
- PMID: 34761294
- PMCID: PMC8776678
- DOI: 10.1007/s10482-021-01676-7
Genome analysis suggests the bacterial family Acetobacteraceae is a source of undiscovered specialized metabolites
Abstract
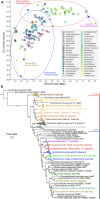
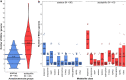
Acetobacteraceae is an economically important family of bacteria that is used for industrial fermentation in the food/feed sector and for the preparation of sorbose and bacterial cellulose. It comprises two major groups: acetous species (acetic acid bacteria) associated with flowers, fruits and insects, and acidophilic species, a phylogenetically basal and physiologically heterogeneous group inhabiting acid or hot springs, sludge, sewage and freshwater environments. Despite the biotechnological importance of the family Acetobacteraceae, the literature does not provide any information about its ability to produce specialized metabolites. We therefore constructed a phylogenomic tree based on concatenated protein sequences from 141 type strains of the family and predicted the presence of small-molecule biosynthetic gene clusters (BGCs) using the antiSMASH tool. This dual approach allowed us to associate certain biosynthetic pathways with particular taxonomic groups. We found that acidophilic and acetous species contain on average ~ 6.3 and ~ 3.4 BGCs per genome, respectively. All the Acetobacteraceae strains encoded proteins involved in hopanoid biosynthesis, with many also featuring genes encoding type-1 and type-3 polyketide and non-ribosomal peptide synthases, and enzymes for aryl polyene, lactone and ribosomal peptide biosynthesis. Our in silico analysis indicated that the family Acetobacteraceae is a potential source of many undiscovered bacterial metabolites and deserves more detailed experimental exploration.
Keywords: Acetobacteraceae; Biosynthesis; Phylogeny; Specialized metabolites.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

