DNA methylation-based classifier and gene expression signatures detect BRCAness in osteosarcoma
- PMID: 34762643
- PMCID: PMC8584788
- DOI: 10.1371/journal.pcbi.1009562
DNA methylation-based classifier and gene expression signatures detect BRCAness in osteosarcoma
Abstract
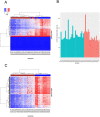
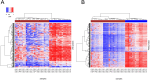
Although osteosarcoma (OS) is a rare cancer, it is the most common primary malignant bone tumor in children and adolescents. BRCAness is a phenotypical trait in tumors with a defect in homologous recombination repair, resembling tumors with inactivation of BRCA1/2, rendering these tumors sensitive to poly (ADP)-ribose polymerase inhibitors (PARPi). Recently, OS was shown to exhibit molecular features of BRCAness. Our goal was to develop a method complementing existing genomic methods to aid clinical decision making on administering PARPi in OS patients. OS samples with DNA-methylation data were divided to BRCAness-positive and negative groups based on the degree of their genomic instability (n = 41). Methylation probes were ranked according to decreasing variance difference between two groups. The top 2000 probes were selected for training and cross-validation of the random forest algorithm. Two-thirds of available OS RNA-Seq samples (n = 17) from the top and bottom of the sample list ranked according to genome instability score were subjected to differential expression and, subsequently, to gene set enrichment analysis (GSEA). The combined accuracy of trained random forest was 85% and the average area under the ROC curve (AUC) was 0.95. There were 449 upregulated and 1,079 downregulated genes in the BRCAness-positive group (fdr < 0.05). GSEA of upregulated genes detected enrichment of DNA replication and mismatch repair and homologous recombination signatures (FWER < 0.05). Validation of the BRCAness classifier with an independent OS set (n = 20) collected later in the course of study showed AUC of 0.87 with an accuracy of 90%. GSEA signatures computed for this test set were matching the ones observed in the training set enrichment analysis. In conclusion, we developed a new classifier based on DNA-methylation patterns that detects BRCAness in OS samples with high accuracy. GSEA identified genome instability signatures. Machine-learning and gene expression approaches add new epigenomic and transcriptomic aspects to already established genomic methods for evaluation of BRCAness in osteosarcoma and can be extended to cancers characterized by genome instability.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

