Siglec-9 defines and restrains a natural killer subpopulation highly cytotoxic to HIV-infected cells
- PMID: 34762717
- PMCID: PMC8584986
- DOI: 10.1371/journal.ppat.1010034
Siglec-9 defines and restrains a natural killer subpopulation highly cytotoxic to HIV-infected cells
Abstract
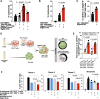
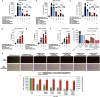
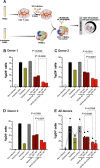
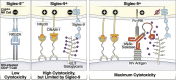
Siglec-9 is an MHC-independent inhibitory receptor expressed on a subset of natural killer (NK) cells. Siglec-9 restrains NK cytotoxicity by binding to sialoglycans (sialic acid-containing glycans) on target cells. Despite the importance of Siglec-9 interactions in tumor immune evasion, their role as an immune evasion mechanism during HIV infection has not been investigated. Using in vivo phenotypic analyses, we found that Siglec-9+ CD56dim NK cells, during HIV infection, exhibit an activated phenotype with higher expression of activating receptors and markers (NKp30, CD38, CD16, DNAM-1, perforin) and lower expression of the inhibitory receptor NKG2A, compared to Siglec-9- CD56dim NK cells. We also found that levels of Siglec-9+ CD56dim NK cells inversely correlate with viral load during viremic infection and CD4+ T cell-associated HIV DNA during suppressed infection. Using in vitro cytotoxicity assays, we confirmed that Siglec-9+ NK cells exhibit higher cytotoxicity towards HIV-infected cells compared to Siglec-9- NK cells. These data are consistent with the notion that Siglec-9+ NK cells are highly cytotoxic against HIV-infected cells. However, blocking Siglec-9 enhanced NK cells' ability to lyse HIV-infected cells, consistent with the known inhibitory function of the Siglec-9 molecule. Together, these data support a model in which the Siglec-9+ CD56dim NK subpopulation is highly cytotoxic against HIV-infected cells even whilst being restrained by the inhibitory effects of Siglec-9. To harness the cytotoxic capacity of the Siglec-9+ NK subpopulation, which is dampened by Siglec-9, we developed a proof-of-concept approach to selectively disrupt Siglec/sialoglycan interactions between NK and HIV-infected cells. We achieved this goal by conjugating Sialidase to several HIV broadly neutralizing antibodies. These conjugates selectively desialylated HIV-infected cells and enhanced NK cells' capacity to kill them. In summary, we identified a novel, glycan-based interaction that may contribute to HIV-infected cells' ability to evade NK immunosurveillance and developed an approach to break this interaction.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, et al.. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 1997;94(24):13193–7. Epub 1997/12/16. doi: 10.1073/pnas.94.24.13193 ; PubMed Central PMCID: PMC24285. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

