Hif-1a suppresses ROS-induced proliferation of cardiac fibroblasts following myocardial infarction
- PMID: 34762860
- PMCID: PMC9021927
- DOI: 10.1016/j.stem.2021.10.009
Hif-1a suppresses ROS-induced proliferation of cardiac fibroblasts following myocardial infarction
Abstract
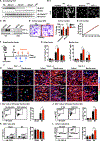
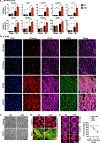
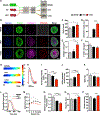
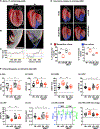
We report that cardiac fibroblasts (CFs) and mesenchymal progenitors are more hypoxic than other cardiac interstitial populations, express more hypoxia-inducible factor 1α (HIF-1α), and exhibit increased glycolytic metabolism. CF-specific deletion of Hif-1a resulted in decreased HIF-1 target gene expression and increased mesenchymal progenitors in uninjured hearts and increased CF activation without proliferation following sham injury, as demonstrated using single-cell RNA sequencing (scRNA-seq). After myocardial infarction (MI), however, there was ∼50% increased CF proliferation and excessive scarring and contractile dysfunction, a scenario replicated in 3D engineered cardiac microtissues. CF proliferation was associated with higher reactive oxygen species (ROS) as occurred also in wild-type mice treated with the mitochondrial ROS generator MitoParaquat (MitoPQ). The mitochondrial-targeted antioxidant MitoTEMPO rescued Hif-1a mutant phenotypes. Thus, HIF-1α in CFs provides a critical braking mechanism against excessive post-ischemic CF activation and proliferation through regulation of mitochondrial ROS. CFs are potential cellular targets for designer antioxidant therapies in cardiovascular disease.
Keywords: 3D cardiac microtissues; Hif-1a; ROS; antioxidant therapies; cardiac fibroblasts; cardiac fibrosis; hypoxia; mesenchymal progenitors; mitochondrial reactive oxygen species; myocardial infarction; single-cell RNA-seq.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Badie N, Satterwhite L, and Bursac N (2009). A method to replicate the microstructure of heart tissue in vitro using DTMRI-based cell micropatterning. Ann. Biomed. Eng 37, 2510–2521. - PubMed
-
- Bergmann O, Zdunek S, Felker A, Salehpour M, Alkass K, Bernard S, Sjostrom SL, Szewczykowska M, Jackowska T, Dos Remedios C, et al. (2015). Dynamics of Cell Generation and Turnover in the Human Heart. Cell 161, 1566–1575. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

