A tissue-bioengineering strategy for modeling rare human kidney diseases in vivo
- PMID: 34764250
- PMCID: PMC8586030
- DOI: 10.1038/s41467-021-26596-y
A tissue-bioengineering strategy for modeling rare human kidney diseases in vivo
Abstract
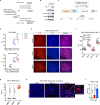
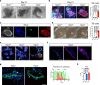
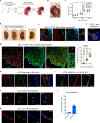
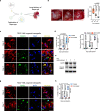
The lack of animal models for some human diseases precludes our understanding of disease mechanisms and our ability to test prospective therapies in vivo. Generation of kidney organoids from Tuberous Sclerosis Complex (TSC) patient-derived-hiPSCs allows us to recapitulate a rare kidney tumor called angiomyolipoma (AML). Organoids derived from TSC2-/- hiPSCs but not from isogenic TSC2+/- or TSC2+/+ hiPSCs share a common transcriptional signature and a myomelanocytic cell phenotype with kidney AMLs, and develop epithelial cysts, replicating two major TSC-associated kidney lesions driven by genetic mechanisms that cannot be consistently recapitulated with transgenic mice. Transplantation of multiple TSC2-/- renal organoids into the kidneys of immunodeficient rats allows us to model AML in vivo for the study of tumor mechanisms, and to test the efficacy of rapamycin-loaded nanoparticles as an approach to rapidly ablate AMLs. Collectively, our experimental approaches represent an innovative and scalable tissue-bioengineering strategy for modeling rare kidney disease in vivo.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Bernstein J, Robbins TO. Renal involvement in tuberous sclerosis. Ann. N. Y Acad. Sci. 1991;615:36–49. - PubMed
-
- Bissler JJ, Christopher Kingswood J. Renal manifestation of tuberous sclerosis complex. Am. J. Med. Genet. Part C., Semin. Med. Genet. 2018;178:338–347. - PubMed
-
- Thway K, Fisher C. PEComa: morphology and genetics of a complex tumor family. Ann. diagnostic Pathol. 2015;19:359–368. - PubMed
-
- Folpe AL, Kwiatkowski DJ. Perivascular epithelioid cell neoplasms: pathology and pathogenesis. Hum. Pathol. 2010;41:1–15. - PubMed
-
- Kaiserling E, Kröber S, Xiao JC, Schaumburg-Lever G. Angiomyolipoma of the kidney. Immunoreactivity with HMB-45 Light- and electron-microscopic findings. Histopathol. 1994;25:41–48. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

