Blocked O-GlcNAc cycling alters mitochondrial morphology, function, and mass
- PMID: 34764359
- PMCID: PMC8586252
- DOI: 10.1038/s41598-021-01512-y
Blocked O-GlcNAc cycling alters mitochondrial morphology, function, and mass
Abstract
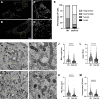
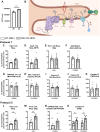
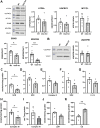
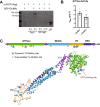
O-GlcNAcylation is a prevalent form of glycosylation that regulates proteins within the cytosol, nucleus, and mitochondria. The O-GlcNAc modification can affect protein cellular localization, function, and signaling interactions. The specific impact of O-GlcNAcylation on mitochondrial morphology and function has been elusive. In this manuscript, the role of O-GlcNAcylation on mitochondrial fission, oxidative phosphorylation (Oxphos), and the activity of electron transport chain (ETC) complexes were evaluated. In a cellular environment with hyper O-GlcNAcylation due to the deletion of O-GlcNAcase (OGA), mitochondria showed a dramatic reduction in size and a corresponding increase in number and total mitochondrial mass. Because of the increased mitochondrial content, OGA knockout cells exhibited comparable coupled mitochondrial Oxphos and ATP levels when compared to WT cells. However, we observed reduced protein levels for complex I and II when comparing normalized mitochondrial content and reduced linked activity for complexes I and III when examining individual ETC complex activities. In assessing mitochondrial fission, we observed increased amounts of O-GlcNAcylated dynamin-related protein 1 (Drp1) in cells genetically null for OGA and in glioblastoma cells. Individual regions of Drp1 were evaluated for O-GlcNAc modifications, and we found that this post-translational modification (PTM) was not limited to the previously characterized residues in the variable domain (VD). Additional modification sites are predicted in the GTPase domain, which may influence enzyme activity. Collectively, these results highlight the impact of O-GlcNAcylation on mitochondrial dynamics and ETC function and mimic the changes that may occur during glucose toxicity from hyperglycemia.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Seo J, Lee KJ. Post-translational modifications and their biological functions: Proteomic analysis and systematic approaches. J. Biochem. Mol. Biol. 2004;37:35–44. - PubMed
-
- Buuh ZY, Lyu Z, Wang RE. Interrogating the roles of post-translational modifications of non-histone proteins. J. Med. Chem. 2018;61:3239–3252. - PubMed
-
- Yang YR, Suh PG. O-GlcNAcylation in cellular functions and human diseases. Adv. Biol. Regul. 2014;54:68–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

