Autocrine vitamin D signaling switches off pro-inflammatory programs of TH1 cells
- PMID: 34764490
- PMCID: PMC7612139
- DOI: 10.1038/s41590-021-01080-3
Autocrine vitamin D signaling switches off pro-inflammatory programs of TH1 cells
Abstract
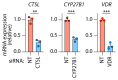
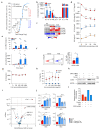
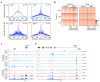
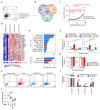
The molecular mechanisms governing orderly shutdown and retraction of CD4+ type 1 helper T (TH1) cell responses remain poorly understood. Here we show that complement triggers contraction of TH1 responses by inducing intrinsic expression of the vitamin D (VitD) receptor and the VitD-activating enzyme CYP27B1, permitting T cells to both activate and respond to VitD. VitD then initiated the transition from pro-inflammatory interferon-γ+ TH1 cells to suppressive interleukin-10+ cells. This process was primed by dynamic changes in the epigenetic landscape of CD4+ T cells, generating super-enhancers and recruiting several transcription factors, notably c-JUN, STAT3 and BACH2, which together with VitD receptor shaped the transcriptional response to VitD. Accordingly, VitD did not induce interleukin-10 expression in cells with dysfunctional BACH2 or STAT3. Bronchoalveolar lavage fluid CD4+ T cells of patients with COVID-19 were TH1-skewed and showed de-repression of genes downregulated by VitD, from either lack of substrate (VitD deficiency) and/or abnormal regulation of this system.
© 2021. This is a U.S. government work and not under copyright protection in the U.S.; foreign copyright protection may apply.
Conflict of interest statement
The authors have no competing interests to declare.
Figures












Comment in
-
Vitamin D shuts down T cell-mediated inflammation.Nat Rev Immunol. 2022 Jan;22(1):1. doi: 10.1038/s41577-021-00663-3. Nat Rev Immunol. 2022. PMID: 34799725 Free PMC article.
References
-
- Gazzinelli RT, et al. In the absence of endogenous IL-10, mice acutely infected with Toxoplasma gondii succumb to a lethal immune response dependent on CD4+ T cells and accompanied by overproduction of IL-12, IFN-gamma and TNF-alpha. J Immunol. 1996;157:798–805. - PubMed
-
- Robbins RA, Russ WD, Rasmussen JK, Clayton MM. Activation of the Complement System in the Adult Respiratory Distress Syndrome1–4. Am Rev Respir Dis. 1987;135:651–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- ZIA AI001175/ImNIH/Intramural NIH HHS/United States
- 097261/Z/11/Z/Wellcome Trust (Wellcome)
- ZIA/Hl006223/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- DK12262401A1/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- R01 DK122624/DK/NIDDK NIH HHS/United States
- ZIA HL006193/ImNIH/Intramural NIH HHS/United States
- FR 3851/2-1/Deutsche Forschungsgemeinschaft (German Research Foundation)
- ZIA/HL006193/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
- ZIA/DK075149/U.S. Department of Health & Human Services | NIH | National Institute of Diabetes and Digestive and Kidney Diseases (National Institute of Diabetes & Digestive & Kidney Diseases)
- ZIA/AI00117/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- RG/13/12/30395/British Heart Foundation (BHF)
- R35 GM138283/GM/NIGMS NIH HHS/United States
- ZIA DK075149/ImNIH/Intramural NIH HHS/United States
- RG/13/12/30395/BHF_/British Heart Foundation/United Kingdom
- CCFA no. 3765/Crohn's and Colitis Foundation (Crohn's & Colitis Foundation)
- R35GM138283/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- WT_/Wellcome Trust/United Kingdom
- ZIA/AI001175/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- ZIA/Hl00622/U.S. Department of Health & Human Services | NIH | National Heart, Lung, and Blood Institute (NHLBI)
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

