ZMYM3 May Promote Cell Proliferation in Small Cell Lung Carcinoma
- PMID: 34764523
- PMCID: PMC8569135
- DOI: 10.1267/ahc.21-00012
ZMYM3 May Promote Cell Proliferation in Small Cell Lung Carcinoma
Abstract
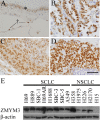
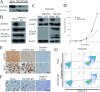
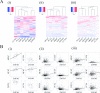
Zinc finger, myeloproliferative, and mental retardation-type containing 3 (ZMYM3) is a highly conserved protein among vertebrates. Although it promotes DNA repair and moderate histone acetylation, the other functions of ZMYM3 remain unclear. We herein examined the physiological functions of ZMYM3 in human lung cancer using a ZMYM3-knockdown small cell lung cancer (SCLC) cell line. ZMYM3-knockdown SCLC cells grew slowly and the Ki-67 labeling index was lower in ZMYM3-knockdown cells than in mock cells. The subcutaneous tumors that formed after xenotransplantation into immunodeficient mice were slightly smaller in the ZMYM3-knockdown group than in the mock group. Furthermore, public RNA-sequencing data analyses showed similar RNA profiles between ZMYM3 and some cell proliferation markers. These results indicate that ZMYM3 promotes cell proliferation in human lung carcinomas, particularly SCLC.
Keywords: ZMYM3; cell proliferation; immunohistochemistry; lung carcinoma; mouse tissues.
2021 The Japan Society of Histochemistry and Cytochemistry.
Conflict of interest statement
VThe authors declare that there are no conflicts of interest.
Figures

References
-
- Alizadeh, F., Bozorgmehr, A., Tavakkoly-Bazzaz, J. and Ohadi, M. (2018) Skewing of the genetic architecture at the ZMYM3 human-specific 5' UTR short tandem repeat in schizophrenia. Mol. Genet. Genomics 293; 747–752. - PubMed
-
- Alizadeh, F., Moharrami, T., Mousavi, N., Yazarlou, F., Bozorgmehr, A., Shahsavand, E., et al. (2019) Disease-only alleles at the extreme ends of the human ZMYM3 exceptionally long 5' UTR short tandem repeat in bipolar disorder: a pilot study. J. Affect. Disord. 251; 86–90. - PubMed

