Deep-learning in situ classification of HIV-1 virion morphology
- PMID: 34765089
- PMCID: PMC8554174
- DOI: 10.1016/j.csbj.2021.10.001
Deep-learning in situ classification of HIV-1 virion morphology
Abstract
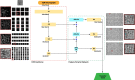
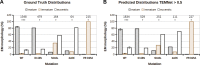
Transmission electron microscopy (TEM) has a multitude of uses in biomedical imaging due to its ability to discern ultrastructure morphology at the nanometer scale. Through its ability to directly visualize virus particles, TEM has for several decades been an invaluable tool in the virologist's toolbox. As applied to HIV-1 research, TEM is critical to evaluate activities of inhibitors that block the maturation and morphogenesis steps of the virus lifecycle. However, both the preparation and analysis of TEM micrographs requires time consuming manual labor. Through the dedicated use of computer vision frameworks and machine learning techniques, we have developed a convolutional neural network backbone of a two-stage Region Based Convolutional Neural Network (RCNN) capable of identifying, segmenting and classifying HIV-1 virions at different stages of maturation and morphogenesis. Our results outperformed common RCNN backbones, achieving 80.0% mean Average Precision on a diverse set of micrographs comprising different experimental samples and magnifications. We expect that this tool will be of interest to a broad range of researchers.
Keywords: Artificial intelligence; Computer vision; Deep learning; Electron microscopy; HIV-1; Quantitative biology; Virology.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures







References
-
- Barre-Sinoussi F., Chermann J., Rey F., Nugeyre M., Chamaret S., Gruest J., Dauguet C., Axler-Blin C., Vezinet-Brun F., Rouzioux C., Rozenbaum W., Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (aids) Science. 1983;220(4599):868–871. doi: 10.1126/science.6189183. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
