COVID-19: insights into virus-receptor interactions
- PMID: 34766003
- PMCID: PMC8035060
- DOI: 10.1186/s43556-021-00033-4
COVID-19: insights into virus-receptor interactions
Abstract
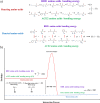
The recent outbreak of Coronavirus Disease 2019 (COVID-19) calls for rapid mobilization of scientists to probe and explore solutions to this deadly disease. A limited understanding of the high transmissibility of SARS-CoV-2 (Severe acute respiratory syndrome coronavirus 2) relative to other coronavirus strains guides a deeper investigation into the virus/receptor interactions. The cutting-edge studies in thermodynamic and kinetic properties of interactions such as protein-protein interplays have been reviewed in many modeling and analysis studies. Highlighting the thermodynamic assessments of biological interactions and emphasizing the boosted transmissibility of SARS-CoV-2 despite its high similarity in structure and sequence with other coronavirus strains is an important and highly valuable investigation that can lead scientists to discover analytical and fundamental approaches in studying virus's interactions. Accordingly, we have attempted to describe the crucial factors such as conformational changes and hydrophobicity particularities that influence on thermodynamic potentials in the SARS-COV-2 S-protein adsorption process. Discussing the thermodynamic potentials and the kinetics of the SARS-CoV-2 S-protein in its interaction with the ACE2 receptors of the host cell is a fundamental approach that would be extremely valuable in designing candidate pharmaceutical agents or exploring alternative treatments.
Keywords: ACE2; COVID-19; Conformational changes; Coronavirus; Glycosylation; Hydrophobicity; SARS-CoV-2; Thermodynamics; Virus-cell interactions.
© The Author(s) 2021.
Conflict of interest statement
Competing interestsThe authors declare no potential conflicts of interests with respect to the research, authorship and/or publication of this article.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous
