A low-cost and open-source protocol to produce key enzymes for molecular detection assays
- PMID: 34766029
- PMCID: PMC8571801
- DOI: 10.1016/j.xpro.2021.100899
A low-cost and open-source protocol to produce key enzymes for molecular detection assays
Abstract
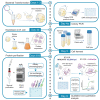
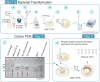
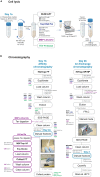
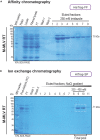
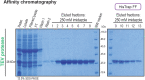
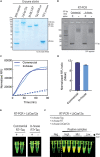
Here, we describe a detailed step-by-step protocol for the expression, purification, quantification, and activity determination of key enzymes for molecular detection of pathogens. Based on previous reports, we optimized the protocol for LbCas12a, Taq DNA polymerase, M-MLV reverse transcriptase, and TEV protease to make it compatible with minimal laboratory equipment, broadly available in low- and middle-income countries. The enzymes produced with this protocol have been successfully used for molecular detection applications. For complete details on the use and execution of this protocol, please refer to Alcántara et al. (2021a, 2021b).
Keywords: Biotechnology and bioengineering; CRISPR; Health Sciences; Microbiology; Molecular Biology; Protein Biochemistry; Protein expression and purification.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Comment in
-
Open access methods and protocols promote open science in a pandemic.STAR Protoc. 2022 Mar 2;3(1):101226. doi: 10.1016/j.xpro.2022.101226. eCollection 2022 Mar 18. STAR Protoc. 2022. PMID: 35284832 Free PMC article.
References
-
- Bradford M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

