Global ubiquitylation analysis of mitochondria in primary neurons identifies endogenous Parkin targets following activation of PINK1
- PMID: 34767452
- PMCID: PMC8589319
- DOI: 10.1126/sciadv.abj0722
Global ubiquitylation analysis of mitochondria in primary neurons identifies endogenous Parkin targets following activation of PINK1
Abstract
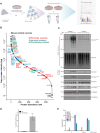
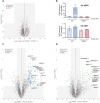
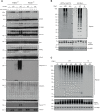
How activation of PINK1 and Parkin leads to elimination of damaged mitochondria by mitophagy is largely based on cell lines with few studies in neurons. Here, we have undertaken proteomic analysis of mitochondria from mouse neurons to identify ubiquitylated substrates of endogenous Parkin. Comparative analysis with human iNeuron datasets revealed a subset of 49 PINK1 activation–dependent diGLY sites in 22 proteins conserved across mouse and human systems. We use reconstitution assays to demonstrate direct ubiquitylation by Parkin in vitro. We also identified a subset of cytoplasmic proteins recruited to mitochondria that undergo PINK1 and Parkin independent ubiquitylation, indicating the presence of alternate ubiquitin E3 ligase pathways that are activated by mitochondrial depolarization in neurons. Last, we have developed an online resource to search for ubiquitin sites and enzymes in mitochondria of neurons, MitoNUb. These findings will aid future studies to understand Parkin activation in neuronal subtypes.
Figures


References
-
- Vafai S. B., Mootha V. K., Mitochondrial disorders as windows into an ancient organelle. Nature 491, 374–383 (2012). - PubMed
-
- Youle R. J., Mitochondria—Striking a balance between host and endosymbiont. Science 365, eaaw9855 (2019). - PubMed
-
- McFarland R., Taylor R. W., Turnbull D. M., A neurological perspective on mitochondrial disease. Lancet Neurol. 9, 829–840 (2010). - PubMed
-
- Montava-Garriga L., Ganley I. G., Outstanding questions in mitophagy: What we do and do not know. J. Mol. Biol. 432, 206–230 (2020). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

