Binding free energy decomposition and multiple unbinding paths of buried ligands in a PreQ1 riboswitch
- PMID: 34767553
- PMCID: PMC8612554
- DOI: 10.1371/journal.pcbi.1009603
Binding free energy decomposition and multiple unbinding paths of buried ligands in a PreQ1 riboswitch
Abstract
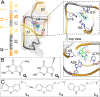
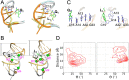
Riboswitches are naturally occurring RNA elements that control bacterial gene expression by binding to specific small molecules. They serve as important models for RNA-small molecule recognition and have also become a novel class of targets for developing antibiotics. Here, we carried out conventional and enhanced-sampling molecular dynamics (MD) simulations, totaling 153.5 μs, to characterize the determinants of binding free energies and unbinding paths for the cognate and synthetic ligands of a PreQ1 riboswitch. Binding free energy analysis showed that two triplets of nucleotides, U6-C15-A29 and G5-G11-C16, contribute the most to the binding of the cognate ligands, by hydrogen bonding and by base stacking, respectively. Mg2+ ions are essential in stabilizing the binding pocket. For the synthetic ligands, the hydrogen-bonding contributions of the U6-C15-A29 triplet are significantly compromised, and the bound state resembles the apo state in several respects, including the disengagement of the C15-A14-A13 and A32-G33 base stacks. The bulkier synthetic ligands lead to significantly loosening of the binding pocket, including extrusion of the C15 nucleobase and a widening of the C15-C30 groove. Enhanced-sampling simulations further revealed that the cognate and synthetic ligands unbind in almost opposite directions. Our work offers new insight for designing riboswitch ligands.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

