Clinical and molecular epidemiology of influenza viruses from Romanian patients hospitalized during the 2019/20 season
- PMID: 34767579
- PMCID: PMC8589178
- DOI: 10.1371/journal.pone.0258798
Clinical and molecular epidemiology of influenza viruses from Romanian patients hospitalized during the 2019/20 season
Abstract
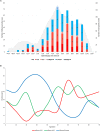
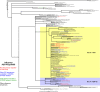
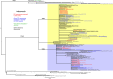
Two main mechanisms contribute to the continuous evolution of influenza viruses: accumulation of mutations in the hemagglutinin and neuraminidase genes (antigenic drift) and genetic re-assortments (antigenic shift). Epidemiological surveillance is important in identifying new genetic variants of influenza viruses with potentially increased pathogenicity and transmissibility. In order to characterize the 2019/20 influenza epidemic in Romania, 1042 respiratory samples were collected from consecutive patients hospitalized with acute respiratory infections in the National Institute for Infectious Diseases "Prof. Dr. Matei Balș", Bucharest Romania and tested for influenza A virus, influenza B virus and respiratory syncytial virus (RSV) by real-time PCR. Out of them, 516 cases were positive for influenza, with relatively equal distribution of influenza A and B. Two patients had influenza A and B co-infection and 8 patients had influenza-RSV co-infection. The most severe cases, requiring supplemental oxygen administration or intensive care, and the most deaths were reported in patients aged 65 years and over. Subtyping showed the predominance of A(H3N2) compared to A(H1N1)pdm09 pdm09 (60.4% and 39.6% of all subtyped influenza A isolates, respectively), and the circulation of Victoria B lineage only. Influenza B started to circulate first (week 47/2019), with influenza A appearing slightly later (week 50/2019), followed by continued co-circulation of A and B viruses throughout the season. Sixty-eight samples, selected to cover the entire influenza season and all circulating viral types, were analysed by next generation sequencing (NGS). All A(H1N1)pdm09 sequences identified during this season in Romania were clustered in the 6b1.A clade (sub-clades: 6b1.A.183P -5a and 6b1.A.187A). For most A(H1N1)pdm09 sequences, the dominant epitope was Sb (pepitope = 0.25), reducing the vaccine efficacy by approximately 60%. According to phylogenetic analysis, influenza A(H3N2) strains circulating in this season belonged predominantly to clade 3C.3A, with only few sequences in clade 3C.2A1b. These 3C.2A1b sequences, two of which belonged to vaccinated patients, harbored mutations in antigenic sites leading to potential reduction of vaccine efficacy. Phylogenetic analysis of influenza B, lineage Victoria, sequences showed that the circulating strains belonged to clade V1A3. As compared to the other viral types, fewer mutations were observed in B/Victoria strains, with limited impact on vaccine efficiency based on estimations.
Conflict of interest statement
OS, Anca S-C, and Adrian S-C report being investigators in influenza clinical trials by Shionogi and F. Hoffmann-La Roche, outside the scope of the submitted work. No other authors have competing interests to declare.
Figures

References
-
- World Health Organization. Recommended composition of influenza virus vaccines for use in the 2021–2022 northern hemisphere influenza season 2021 [cited 2021, Apr 9]]. Available from: https://www.who.int/publications/i/item/recommended-composition-of-influ....
-
- Suntronwong N, Klinfueng S, Korkong S, Vichaiwattana P, Thongmee T, Vongpunsawad S, et al. Characterizing genetic and antigenic divergence from vaccine strain of influenza A and B viruses circulating in Thailand, 2017–2020. Sci Rep. 2021;11(1):735. Epub 2021/01/14. doi: 10.1038/s41598-020-80895-w - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

