Enzymatic Cβ-H Functionalization of l-Arg and l-Leu in Nonribosomally Derived Peptidyl Natural Products: A Tale of Two Oxidoreductases
- PMID: 34767710
- PMCID: PMC10258837
- DOI: 10.1021/jacs.1c08177
Enzymatic Cβ-H Functionalization of l-Arg and l-Leu in Nonribosomally Derived Peptidyl Natural Products: A Tale of Two Oxidoreductases
Abstract
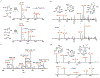
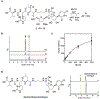
Muraymycins are peptidyl nucleoside antibiotics that contain two Cβ-modified amino acids, (2S,3S)-capreomycidine and (2S,3S)-β-OH-Leu. The former is also a component of chymostatins, which are aldehyde-containing peptidic protease inhibitors that─like muraymycin─are derived from nonribosomal peptide synthetases (NRPSs). Using feeding experiments and in vitro characterization of 12 recombinant proteins, the biosynthetic mechanism for both nonproteinogenic amino acids is now defined. The formation of (2S,3S)-capreomycidine is shown to involve an FAD-dependent dehydrogenase:cyclase that requires an NRPS-bound pathway intermediate as a substrate. This cryptic dehydrogenation strategy is both temporally and mechanistically distinct in comparison to the biosynthesis of other capreomycidine diastereomers, which has previously been shown to proceed by Cβ-hydroxylation of free l-Arg catalyzed by a member of the nonheme Fe2+- and α-ketoglutarate (αKG)-dependent dioxygenase family and (eventually) a dehydration-mediated cyclization process catalyzed by a distinct enzyme(s). Contrary to our initial expectation, the sole nonheme Fe2+- and αKG-dependent dioxygenase candidate Mur15 encoded within the muraymycin gene cluster is instead demonstrated to catalyze specific Cβ hydroxylation of the Leu residue to generate (2S,3S)-β-OH-Leu that is found in most muraymycin congeners. Importantly, and in contrast to known l-Arg-Cβ-hydroxylases, the Mur15-catalyzed reaction occurs after the NRPS-mediated assembly of the peptide scaffold. This late-stage functionalization affords the opportunity to exploit Mur15 as a biocatalyst, proof of concept of which is provided.
Conflict of interest statement
CONFLICT OF INTEREST DISCLOSURE
Z. C. and S. V. L. are Inventors on a provisional patent application for Mur15 biocatalysis, filed through the University of Kentucky Research Foundation
Figures

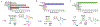

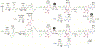
Similar articles
-
Biosynthesis of Glidomides and Elucidation of Different Mechanisms for Formation of β-OH Amino Acid Building Blocks.Angew Chem Int Ed Engl. 2022 Aug 26;61(35):e202203591. doi: 10.1002/anie.202203591. Epub 2022 Jun 29. Angew Chem Int Ed Engl. 2022. PMID: 35689369
-
Characterization and Structural Determination of CmnG-A, the Adenylation Domain That Activates the Nonproteinogenic Amino Acid Capreomycidine in Capreomycin Biosynthesis.Chembiochem. 2022 Dec 16;23(24):e202200563. doi: 10.1002/cbic.202200563. Epub 2022 Nov 16. Chembiochem. 2022. PMID: 36278314
-
Structural basis of nonribosomal peptide macrocyclization in fungi.Nat Chem Biol. 2016 Dec;12(12):1001-1003. doi: 10.1038/nchembio.2202. Epub 2016 Oct 17. Nat Chem Biol. 2016. PMID: 27748753 Free PMC article.
-
Structure and noncanonical chemistry of nonribosomal peptide biosynthetic machinery.Nat Prod Rep. 2012 Oct;29(10):1099-110. doi: 10.1039/c2np20023f. Epub 2012 Jun 25. Nat Prod Rep. 2012. PMID: 22729219 Free PMC article. Review.
-
Chemical Strategies for Visualizing and Analyzing Endogenous Nonribosomal Peptide Synthetase (NRPS) Megasynthetases.Chembiochem. 2019 Aug 16;20(16):2032-2040. doi: 10.1002/cbic.201900186. Epub 2019 Jul 22. Chembiochem. 2019. PMID: 31134733 Review.
Cited by
-
Mechanistic and Structural Insights into a Divergent PLP-Dependent l-Enduracididine Cyclase from a Toxic Cyanobacterium.ACS Catal. 2023 Jul 11;13(14):9817-9828. doi: 10.1021/acscatal.3c01294. eCollection 2023 Jul 21. ACS Catal. 2023. PMID: 37497377 Free PMC article.
-
Mechanistic and structural insights into a divergent PLP-dependent L-enduracididine cyclase from a toxic cyanobacterium.bioRxiv [Preprint]. 2023 Mar 21:2023.03.21.533663. doi: 10.1101/2023.03.21.533663. bioRxiv. 2023. Update in: ACS Catal. 2023 Jul 11;13(14):9817-9828. doi: 10.1021/acscatal.3c01294. PMID: 36993528 Free PMC article. Updated. Preprint.
-
PEARL-Catalyzed Peptide Bond Formation after Chain Reversal by Ureido-Forming Condensation Domains.ACS Cent Sci. 2024 Jun 3;10(6):1242-1250. doi: 10.1021/acscentsci.4c00044. eCollection 2024 Jun 26. ACS Cent Sci. 2024. PMID: 38947204 Free PMC article.
-
Promiscuous Enzymes for Residue-Specific Peptide and Protein Late-Stage Functionalization.Chembiochem. 2023 Sep 1;24(17):e202300372. doi: 10.1002/cbic.202300372. Epub 2023 Jul 19. Chembiochem. 2023. PMID: 37338668 Free PMC article. Review.
References
-
- Lorion MM; Kaplaneris N; Son J; Kuniyil R; Lutz A Late-stage peptide diversification through cobalt-catalyzed C-H activation: Sequential multicatalysis for stapled peptides. Angew. Chem. Int. Ed 2019, 58, 1684–1688. - PubMed
-
- Sengupta S; Mehta G Late stage modification of peptides via C-H activation reactions. Tetrahedron Lett. 2017, 58, 1357–1372.
-
- Carter GT; Lotvin JA; McDonald LA U.S. Patent 20030104986A1, June 5, 2003.
-
- McDonald LA; Barbieri LR; Carter GT; Lenoy E; Lotvin J; Peterson PJ; Siegel MM; Singh G; Williamson RT Structures of the muraymycins, novel peptidoglycan biosynthesis inhibitors. J. Am. Chem. Soc 2002, 124, 10260–10261. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

