Inhibitory monoclonal antibody targeting ADAM17 expressed on cancer cells
- PMID: 34768098
- PMCID: PMC8592942
- DOI: 10.1016/j.tranon.2021.101265
Inhibitory monoclonal antibody targeting ADAM17 expressed on cancer cells
Abstract
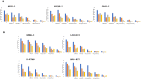
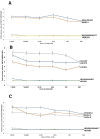
ADAM17 is upregulated in many cancers and in turn activates signaling pathways, including EGFR/ErbB, as well as those underlying resistance to targeted anti-EGFR therapies. Due to its central role in oncogenic pathways and drug resistance mechanisms, specific and efficacious monoclonal antibodies against ADAM17 could be useful for a broad patient population with solid tumors. Hence, we describe here an inhibitory anti-ADAM17 monoclonal antibody, named D8P1C1, that preferentially recognizes ADAM17 on cancer cells. D8P1C1 inhibits the catalytic activity of ADAM17 in a fluorescence-based peptide cleavage assay, as well as the proliferation of a range of cancer cell lines, including breast, ovarian, glioma, colon and the lung adenocarcinoma. In mouse models of triple-negative breast cancer and ovarian cancer, treatment with the mAb results in 78% and 45% tumor growth inhibition, respectively. Negative staining electron microscopy analysis of the ADAM17 ectodomain in complex with D8P1C1 reveals that the mAb binds the ADAM17 protease domain, consistent with its ability to inhibit the ADAM17 catalytic activity. Collectively, our results demonstrate the therapeutic potential of the D8P1C1 mAb to treat solid tumors.
Keywords: ADAM17; Cancer therapy; EGFR signaling; Monoclonal antibody.
Copyright © 2021. Published by Elsevier Inc.
Conflict of interest statement
The authors declare that they have no competing interest.
Figures
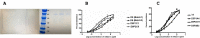



References
-
- Murphy G. The ADAMS: signalling scissors in the tumour microenvironment. Nat. Rev. Cancer. 2008;8:929–941. - PubMed
-
- Dreymueller D., Uhlig S., Ludwig A. ADAM-family metalloproteinases in lung inflammation: potential therapeutic targets. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2015;308(4):L325–L343. - PubMed
-
- Pruessmeyer J., Ludwig A. The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin. Cell Dev. Biol. 2009;20(2):164–174. - PubMed
-
- Seals D.F., Courtneidge SA S.A. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17(1):7–30. - PubMed
-
- Janes P.W., Saha N., Barton W.A., Kolev M.V., Wimmer-Kleikamp S.H., Nievergall E., Blobel C.P., Himanen J.P., Lackmann M., Nikolov D.B. Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans. Cell. 2005;123(2):291–304. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

