Inducible Enrichment of Osa-miR1432 Confers Rice Bacterial Blight Resistance through Suppressing OsCaML2
- PMID: 34768797
- PMCID: PMC8583624
- DOI: 10.3390/ijms222111367
Inducible Enrichment of Osa-miR1432 Confers Rice Bacterial Blight Resistance through Suppressing OsCaML2
Abstract
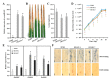
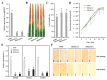
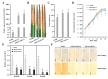
MicroRNAs (miRNAs) handle immune response to pathogens by adjusting the function of target genes in plants. However, the experimentally documented miRNA/target modules implicated in the interplay between rice and Xanthomonas oryzae pv. oryzae (Xoo) are still in the early stages. Herein, the expression of osa-miR1432 was induced in resistant genotype IRBB5, but not susceptible genotype IR24, under Xoo strain PXO86 attack. Overexpressed osa-miR1432 heightened rice disease resistance to Xoo, indicated by enhancive enrichment of defense marker genes, raised reactive oxygen species (ROS) levels, repressed bacterial growth and shortened leaf lesion length, whilst the disruptive accumulation of osa-miR1432 accelerated rice susceptibility to Xoo infection. Noticeably, OsCaML2 (LOC_Os03g59770) was experimentally confirmed as a target gene of osa-miR1432, and the overexpressing OsCaML2 transgenic plants exhibited compromised resistance to Xoo infestation. Our results indicate that osa-miR1432 and OsCaML2 were differently responsive to Xoo invasion at the transcriptional level and fine-tune rice resistance to Xoo infection, which may be referable in resistance gene discovery and valuable in the pursuit of improving Xoo resistance in rice breeding.
Keywords: OsCaML2; osa-miR1432; rice bacterial blight.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
The Roles of MicroRNAs in the Regulation of Rice-Pathogen Interactions.Plants (Basel). 2025 Jan 6;14(1):136. doi: 10.3390/plants14010136. Plants (Basel). 2025. PMID: 39795396 Free PMC article. Review.
-
Characteristic Dissection of Xanthomonas oryzae pv. oryzae Responsive MicroRNAs in Rice.Int J Mol Sci. 2020 Jan 25;21(3):785. doi: 10.3390/ijms21030785. Int J Mol Sci. 2020. PMID: 31991765 Free PMC article.
-
A Rice NBS-ARC Gene Conferring Quantitative Resistance to Bacterial Blight Is Regulated by a Pathogen Effector-Inducible miRNA.Mol Plant. 2020 Dec 7;13(12):1752-1767. doi: 10.1016/j.molp.2020.09.015. Epub 2020 Sep 20. Mol Plant. 2020. PMID: 32966899
-
Identification and characterization of genes frequently responsive to Xanthomonas oryzae pv. oryzae and Magnaporthe oryzae infections in rice.BMC Genomics. 2020 Jan 6;21(1):21. doi: 10.1186/s12864-019-6438-y. BMC Genomics. 2020. PMID: 31906847 Free PMC article.
-
MicroRNA-mediated host defense mechanisms against pathogens and herbivores in rice: balancing gains from genetic resistance with trade-offs to productivity potential.BMC Plant Biol. 2022 Jul 18;22(1):351. doi: 10.1186/s12870-022-03723-5. BMC Plant Biol. 2022. PMID: 35850632 Free PMC article. Review.
Cited by
-
The Roles of MicroRNAs in the Regulation of Rice-Pathogen Interactions.Plants (Basel). 2025 Jan 6;14(1):136. doi: 10.3390/plants14010136. Plants (Basel). 2025. PMID: 39795396 Free PMC article. Review.
-
Genome-wide analysis of the peanut CaM/CML gene family reveals that the AhCML69 gene is associated with resistance to Ralstonia solanacearum.BMC Genomics. 2024 Feb 21;25(1):200. doi: 10.1186/s12864-024-10108-5. BMC Genomics. 2024. PMID: 38378471 Free PMC article.
-
Genome-Wide Characterization of CaM/CML Gene Family in Cabbage (Brassica oleracea var. capitata): Expression Profiling and Functional Implications During Hyaloperonospora parasitica Infection.Int J Mol Sci. 2025 Mar 30;26(7):3208. doi: 10.3390/ijms26073208. Int J Mol Sci. 2025. PMID: 40244053 Free PMC article.
-
Transcriptome Analysis Reveals the Role of OsCBM1 in Rice Defense Against Xanthomonas oryzae pv.oryzae.Biomolecules. 2025 Feb 14;15(2):287. doi: 10.3390/biom15020287. Biomolecules. 2025. PMID: 40001590 Free PMC article.
-
Mechanism of Rice Resistance to Bacterial Leaf Blight via Phytohormones.Plants (Basel). 2024 Sep 10;13(18):2541. doi: 10.3390/plants13182541. Plants (Basel). 2024. PMID: 39339516 Free PMC article. Review.
References
-
- Lee B.M., Park Y.J., Park D.S., Kang H.W., Kim J.G., Song E.S., Park I.C., Yoon U.H., Hahn J.H., Koo B.S., et al. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 2005;33:577–586. doi: 10.1093/nar/gki206. - DOI - PMC - PubMed
-
- Jambhulkar P.P., Sharma P. Developent of bioformulation and delivery system of Pseudomonas fluorescens against bacterial leaf blight of rice (Xanthomonas oryzae pv. oryzae). J. Environ. Biol. 2014;35:843–849. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

