PRC1 Stabilizes Cardiac Contraction by Regulating Cardiac Sarcomere Assembly and Cardiac Conduction System Construction
- PMID: 34768802
- PMCID: PMC8583368
- DOI: 10.3390/ijms222111368
PRC1 Stabilizes Cardiac Contraction by Regulating Cardiac Sarcomere Assembly and Cardiac Conduction System Construction
Abstract
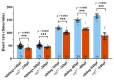
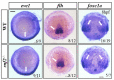
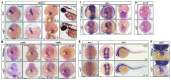
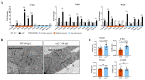
Cardiac development is a complex process that is strictly controlled by various factors, including PcG protein complexes. Several studies have reported the critical role of PRC2 in cardiogenesis. However, little is known about the regulation mechanism of PRC1 in embryonic heart development. To gain more insight into the mechanistic role of PRC1 in cardiogenesis, we generated a PRC1 loss-of-function zebrafish line by using the CRISPR/Cas9 system targeting rnf2, a gene encoding the core subunit shared by all PRC1 subfamilies. Our results revealed that Rnf2 is not involved in cardiomyocyte differentiation and heart tube formation, but that it is crucial to maintaining regular cardiac contraction. Further analysis suggested that Rnf2 loss-of-function disrupted cardiac sarcomere assembly through the ectopic activation of non-cardiac sarcomere genes in the developing heart. Meanwhile, Rnf2 deficiency disrupts the construction of the atrioventricular canal and the sinoatrial node by modulating the expression of bmp4 and other atrioventricular canal marker genes, leading to an impaired cardiac conduction system. The disorganized cardiac sarcomere and defective cardiac conduction system together contribute to defective cardiac contraction. Our results emphasize the critical role of PRC1 in the cardiac development.
Keywords: PRC1; Rnf2; cardiac conduction system; cardiac contraction; sarcomere assembly.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

