Functional Amino Acids and Autophagy: Diverse Signal Transduction and Application
- PMID: 34768858
- PMCID: PMC8592284
- DOI: 10.3390/ijms222111427
Functional Amino Acids and Autophagy: Diverse Signal Transduction and Application
Abstract
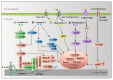
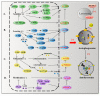
Functional amino acids provide great potential for treating autophagy-related diseases by regulating autophagy. The purpose of the autophagy process is to remove unwanted cellular contents and to recycle nutrients, which is controlled by many factors. Disordered autophagy has been reported to be associated with various diseases, such as cancer, neurodegeneration, aging, and obesity. Autophagy cannot be directly controlled and dynamic amino acid levels are sufficient to regulate autophagy. To date, arginine, leucine, glutamine, and methionine are widely reported functional amino acids that regulate autophagy. As a signal relay station, mammalian target of rapamycin complex 1 (mTORC1) turns various amino acid signals into autophagy signaling pathways for functional amino acids. Deficiency or supplementation of functional amino acids can immediately regulate autophagy and is associated with autophagy-related disease. This review summarizes the mechanisms currently involved in autophagy and amino acid sensing, diverse signal transduction among functional amino acids and autophagy, and the therapeutic appeal of amino acids to autophagy-related diseases. We aim to provide a comprehensive overview of the mechanisms of amino acid regulation of autophagy and the role of functional amino acids in clinical autophagy-related diseases and to further convert these mechanisms into feasible therapeutic applications.
Keywords: autophagy; autophagy-related diseases; functional amino acids; mTORC1; signal transduction.
Conflict of interest statement
The authors declare that they have no competing interest.
Figures


Similar articles
-
AMPK Inhibits ULK1-Dependent Autophagosome Formation and Lysosomal Acidification via Distinct Mechanisms.Mol Cell Biol. 2018 Apr 30;38(10):e00023-18. doi: 10.1128/MCB.00023-18. Print 2018 May 15. Mol Cell Biol. 2018. PMID: 29507183 Free PMC article.
-
Glutamine and asparagine activate mTORC1 independently of Rag GTPases.J Biol Chem. 2020 Mar 6;295(10):2890-2899. doi: 10.1074/jbc.AC119.011578. Epub 2020 Feb 4. J Biol Chem. 2020. PMID: 32019866 Free PMC article.
-
Glutamine metabolism regulates autophagy-dependent mTORC1 reactivation during amino acid starvation.Nat Commun. 2017 Aug 24;8(1):338. doi: 10.1038/s41467-017-00369-y. Nat Commun. 2017. PMID: 28835610 Free PMC article.
-
Amino acid-dependent control of mTORC1 signaling: a variety of regulatory modes.J Biomed Sci. 2020 Aug 17;27(1):87. doi: 10.1186/s12929-020-00679-2. J Biomed Sci. 2020. PMID: 32799865 Free PMC article. Review.
-
mTORC1 as the main gateway to autophagy.Essays Biochem. 2017 Dec 12;61(6):565-584. doi: 10.1042/EBC20170027. Print 2017 Dec 12. Essays Biochem. 2017. PMID: 29233869 Free PMC article. Review.
Cited by
-
Relationship Between Dietary Nutrient Intake and Autophagy-Related Genes in Obese Humans: A Narrative Review.Nutrients. 2024 Nov 22;16(23):4003. doi: 10.3390/nu16234003. Nutrients. 2024. PMID: 39683397 Free PMC article. Review.
-
Duality of Branched-Chain Amino Acids in Chronic Cardiovascular Disease: Potential Biomarkers versus Active Pathophysiological Promoters.Nutrients. 2024 Jun 20;16(12):1972. doi: 10.3390/nu16121972. Nutrients. 2024. PMID: 38931325 Free PMC article. Review.
-
Dysregulation of acyl carnitines, pentose phosphate pathway and arginine and ornithine metabolism are associated with decline in intrinsic capacity in Chinese older adults.Aging Clin Exp Res. 2024 Feb 12;36(1):36. doi: 10.1007/s40520-023-02654-x. Aging Clin Exp Res. 2024. PMID: 38345670 Free PMC article.
-
Autophagy: Are Amino Acid Signals Dependent on the mTORC1 Pathway or Independent?Curr Issues Mol Biol. 2024 Aug 13;46(8):8780-8793. doi: 10.3390/cimb46080519. Curr Issues Mol Biol. 2024. PMID: 39194736 Free PMC article. Review.
-
Amino Acid Metabolism and Autophagy in Atherosclerotic Cardiovascular Disease.Biomolecules. 2024 Dec 6;14(12):1557. doi: 10.3390/biom14121557. Biomolecules. 2024. PMID: 39766264 Free PMC article. Review.
References
-
- Radewa J. Observations on autophagocytosis phenomena in the blood. Z. Rheumaforsch. 1963;22:36–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

