Ensemble of Template-Free and Template-Based Classifiers for Protein Secondary Structure Prediction
- PMID: 34768880
- PMCID: PMC8583764
- DOI: 10.3390/ijms222111449
Ensemble of Template-Free and Template-Based Classifiers for Protein Secondary Structure Prediction
Abstract
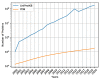
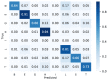
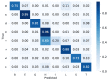
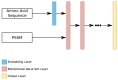
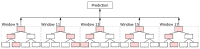
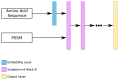
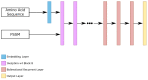
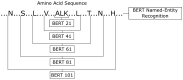
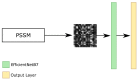
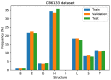
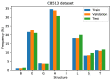
Protein secondary structures are important in many biological processes and applications. Due to advances in sequencing methods, there are many proteins sequenced, but fewer proteins with secondary structures defined by laboratory methods. With the development of computer technology, computational methods have (started to) become the most important methodologies for predicting secondary structures. We evaluated two different approaches to this problem-driven by the recent results obtained by computational methods in this task-(i) template-free classifiers, based on machine learning techniques; and (ii) template-based classifiers, based on searching tools. Both approaches are formed by different sub-classifiers-six for template-free and two for template-based, each with a specific view of the protein. Our results show that these ensembles improve the results of each approach individually.
Keywords: BLAST; deep learning; ensemble; machine learning; protein secondary structure prediction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Protein Secondary Structure Prediction Based on Data Partition and Semi-Random Subspace Method.Sci Rep. 2018 Jun 29;8(1):9856. doi: 10.1038/s41598-018-28084-8. Sci Rep. 2018. PMID: 29959372 Free PMC article.
-
Protein tertiary structure modeling driven by deep learning and contact distance prediction in CASP13.Proteins. 2019 Dec;87(12):1165-1178. doi: 10.1002/prot.25697. Epub 2019 Apr 25. Proteins. 2019. PMID: 30985027 Free PMC article.
-
Recent Advances in Computational Prediction of Secondary and Supersecondary Structures from Protein Sequences.Methods Mol Biol. 2025;2870:1-19. doi: 10.1007/978-1-0716-4213-9_1. Methods Mol Biol. 2025. PMID: 39543027 Review.
-
A hybrid genetic-neural system for predicting protein secondary structure.BMC Bioinformatics. 2005 Dec 1;6 Suppl 4(Suppl 4):S3. doi: 10.1186/1471-2105-6-S4-S3. BMC Bioinformatics. 2005. PMID: 16351752 Free PMC article.
-
Reviewing ensemble classification methods in breast cancer.Comput Methods Programs Biomed. 2019 Aug;177:89-112. doi: 10.1016/j.cmpb.2019.05.019. Epub 2019 May 20. Comput Methods Programs Biomed. 2019. PMID: 31319964 Review.
Cited by
-
TEMPROT: protein function annotation using transformers embeddings and homology search.BMC Bioinformatics. 2023 Jun 8;24(1):242. doi: 10.1186/s12859-023-05375-0. BMC Bioinformatics. 2023. PMID: 37291492 Free PMC article.
-
Protein structure prediction via deep learning: an in-depth review.Front Pharmacol. 2025 Apr 3;16:1498662. doi: 10.3389/fphar.2025.1498662. eCollection 2025. Front Pharmacol. 2025. PMID: 40248099 Free PMC article. Review.
References
-
- Kumar P., Bankapur S., Patil N. An Enhanced Protein Secondary Structure Prediction using Deep Learning Framework on Hybrid Profile based Features. Appl. Soft Comput. 2020;86:105926. doi: 10.1016/j.asoc.2019.105926. - DOI
-
- Oliveira G.B., Pedrini H., Dias Z. Ensemble of Bidirectional Recurrent Networks and Random Forests for Protein Secondary Structure Prediction; Proceedings of the 27th International Conference on Systems, Signals and Image Processing (IWSSIP); Rio de Janeiro, Brazil. 1–3 July 2020; pp. 311–316.
-
- Oliveira G.B., Pedrini H., Dias Z. Protein Secondary Structure Prediction Based on Fusion of Machine Learning Classifiers; Proceedings of the 36th ACM/SIGAPP Symposium On Applied Computing—Bioinformatics Track (ACM SAC BIO); Gwangju, Korea. 22–26 March 2021; pp. 26–29.
-
- Cheng J., Liu Y., Ma Y. Protein Secondary Structure Prediction based on Integration of CNN and LSTM Model. J. Vis. Commun. Image Represent. 2020;71:102844. doi: 10.1016/j.jvcir.2020.102844. - DOI
-
- Cerri R., Mantovani R.G., Basgalupp M.P., de Carvalho A.C. Multi-label Feature Selection Techniques for Hierarchical Multi-label Protein Function Prediction; Proceedings of the International Joint Conference on Neural Networks (IJCNN); Rio de Janeiro, Brazil. 8–13 July 2018; pp. 1–7.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

