Transient Fluorescence Labeling: Low Affinity-High Benefits
- PMID: 34769228
- PMCID: PMC8583718
- DOI: 10.3390/ijms222111799
Transient Fluorescence Labeling: Low Affinity-High Benefits
Abstract
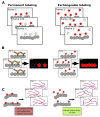
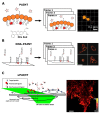
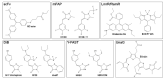
Fluorescent labeling is an established method for visualizing cellular structures and dynamics. The fundamental diffraction limit in image resolution was recently bypassed with the development of super-resolution microscopy. Notably, both localization microscopy and stimulated emission depletion (STED) microscopy impose tight restrictions on the physico-chemical properties of labels. One of them-the requirement for high photostability-can be satisfied by transiently interacting labels: a constant supply of transient labels from a medium replenishes the loss in the signal caused by photobleaching. Moreover, exchangeable tags are less likely to hinder the intrinsic dynamics and cellular functions of labeled molecules. Low-affinity labels may be used both for fixed and living cells in a range of nanoscopy modalities. Nevertheless, the design of optimal labeling and imaging protocols with these novel tags remains tricky. In this review, we highlight the pros and cons of a wide variety of transiently interacting labels. We further discuss the state of the art and future perspectives of low-affinity labeling methods.
Keywords: PAINT; exchangeable labels; fluorescent labeling; super-resolution microscopy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

