Fibroblast Differentiation and Matrix Remodeling Impaired under Simulated Microgravity in 3D Cell Culture Model
- PMID: 34769342
- PMCID: PMC8584780
- DOI: 10.3390/ijms222111911
Fibroblast Differentiation and Matrix Remodeling Impaired under Simulated Microgravity in 3D Cell Culture Model
Abstract
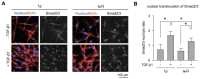
Exposure to microgravity affects astronauts' health in adverse ways. However, less is known about the extent to which fibroblast differentiation during the wound healing process is affected by the lack of gravity. One of the key steps of this process is the differentiation of fibroblasts into myofibroblasts, which contribute functionally through extracellular matrix production and remodeling. In this work, we utilized collagen-based three-dimensional (3D) matrices to mimic interstitial tissue and studied fibroblast differentiation under simulated microgravity (sµG). Our results demonstrated that alpha-smooth muscle actin (αSMA) expression and translocation of Smad2/3 into the cell nucleus were reduced upon exposure to sµG compared to the 1g control, which suggests the impairment of fibroblast differentiation under sµG. Moreover, matrix remodeling and production were decreased under sµG, which is in line with the impaired fibroblast differentiation. We further investigated changes on a transcriptomic level using RNA sequencing. The results demonstrated that sµG has less effect on fibroblast transcriptomes, while sµG triggers changes in the transcriptome of myofibroblasts. Several genes and biological pathways found through transcriptome analysis have previously been reported to impair fibroblast differentiation. Overall, our data indicated that fibroblast differentiation, as well as matrix production and remodeling, are impaired in 3D culture under sµG conditions.
Keywords: 3D cell culture; fibroblast differentiation; matrix remodeling; microgravity; tissue repair.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Monici M., Cialdai F., Romano G., Fusi F., Egli M., Pezzatini S., Morbidelli L. An in Vitro Study on Tissue Repair: Impact of Unloading on Cells Involved in the Remodelling Phase. Microgravity Sci. Technol. 2011;23:391–401. doi: 10.1007/s12217-011-9259-4. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

