A Review of Human Coronaviruses' Receptors: The Host-Cell Targets for the Crown Bearing Viruses
- PMID: 34770863
- PMCID: PMC8587140
- DOI: 10.3390/molecules26216455
A Review of Human Coronaviruses' Receptors: The Host-Cell Targets for the Crown Bearing Viruses
Abstract
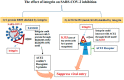
A novel human coronavirus prompted considerable worry at the end of the year 2019. Now, it represents a significant global health and economic burden. The newly emerged coronavirus disease caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is the primary reason for the COVID-19 global pandemic. According to recent global figures, COVID-19 has caused approximately 243.3 million illnesses and 4.9 million deaths. Several human cell receptors are involved in the virus identification of the host cells and entering them. Hence, understanding how the virus binds to host-cell receptors is crucial for developing antiviral treatments and vaccines. The current work aimed to determine the multiple host-cell receptors that bind with SARS-CoV-2 and other human coronaviruses for the purpose of cell entry. Extensive research is needed using neutralizing antibodies, natural chemicals, and therapeutic peptides to target those host-cell receptors in extremely susceptible individuals. More research is needed to map SARS-CoV-2 cell entry pathways in order to identify potential viral inhibitors.
Keywords: COVID-19; SARS-CoV-2; cell receptor; human coronavirus; spike; viral entry.
Conflict of interest statement
The authors declare that there are no conflict of interest.
Figures


References
-
- Payne S. Family Coronaviridae. Viruses. 2017:149–158. doi: 10.1016/B978-0-12-803109-4.00017-9. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

