Selection and Characterization of Vimentin-Binding Aptamer Motifs for Ovarian Cancer
- PMID: 34770931
- PMCID: PMC8588432
- DOI: 10.3390/molecules26216525
Selection and Characterization of Vimentin-Binding Aptamer Motifs for Ovarian Cancer
Abstract
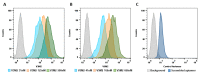
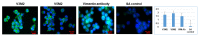
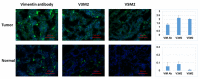
The application of aptamers in biomedicine is emerging as an essential technology in the field of cancer research. As small single-stranded DNA or RNA ligands with high specificity and low immunogenicity for their targets, aptamers provide many advantages in cancer therapeutics over protein-based molecules, such as antibodies. Vimentin is an intermediate filament protein that is overexpressed in endothelial cells of cancerous tissue. High expression levels of vimentin have been associated with increased capacity for migration and invasion of the tumor cells. We have selected and identified thioated aptamers with high specificity for vimentin using human ovarian cancer tissues. Tentative binding motifs were chosen for two vimentin aptamers based on predicted secondary structures. Each of these shorter, tentative binding motifs was synthesized, purified, and characterized via cell binding assays. Two vimentin binding motifs with high fidelity binding were selected and further characterized via cell and tissue binding assays, as well as flow cytometric analysis. The equilibrium binding constants of these small thioated aptamer constructs were also determined. Future applications for the vimentin binding aptamer motifs include conjugation of the aptamers to synthetic dyes for use in targeted imaging and therapy, and ultimately more detailed and precise monitoring of treatment response and tumor progression in ovarian pathology.
Keywords: aptamer; binding motifs; ovarian cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Dyke C.K., Steinhubl S.R., Kleiman N.S., Cannon R.O., Aberle L.G., Lin M., Myles S.K., Melloni C., Harrington R.A., Alexander J.H., et al. First-in-human experience of an antidote-controlled anticoagulant using RNA aptamer technology: A phase 1a pharmacodynamic evaluation of a drug-antidote pair for the controlled regulation of factor IXa activity. Circulation. 2006;114:2490–2497. doi: 10.1161/CIRCULATIONAHA.106.668434. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

