Robust Saliva-Based RNA Extraction-Free One-Step Nucleic Acid Amplification Test for Mass SARS-CoV-2 Monitoring
- PMID: 34771026
- PMCID: PMC8588466
- DOI: 10.3390/molecules26216617
Robust Saliva-Based RNA Extraction-Free One-Step Nucleic Acid Amplification Test for Mass SARS-CoV-2 Monitoring
Abstract
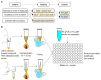
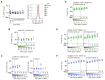
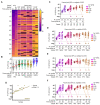
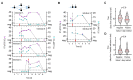
Early diagnosis with rapid detection of the virus plays a key role in preventing the spread of infection and in treating patients effectively. In order to address the need for a straightforward detection of SARS-CoV-2 infection and assessment of viral spread, we developed rapid, sensitive, extraction-free one-step reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and reverse transcription loop-mediated isothermal amplification (RT-LAMP) tests for detecting SARS-CoV-2 in saliva. We analyzed over 700 matched pairs of saliva and nasopharyngeal swab (NSB) specimens from asymptomatic and symptomatic individuals. Saliva, as either an oral cavity swab or passive drool, was collected in an RNA stabilization buffer. The stabilized saliva specimens were heat-treated and directly analyzed without RNA extraction. The diagnostic sensitivity of saliva-based RT-qPCR was at least 95% in individuals with subclinical infection and outperformed RT-LAMP, which had at least 70% sensitivity when compared to NSBs analyzed with a clinical RT-qPCR test. The diagnostic sensitivity for passive drool saliva was higher than that of oral cavity swab specimens (95% and 87%, respectively). A rapid, sensitive one-step extraction-free RT-qPCR test for detecting SARS-CoV-2 in passive drool saliva is operationally simple and can be easily implemented using existing testing sites, thus allowing high-throughput, rapid, and repeated testing of large populations. Furthermore, saliva testing is adequate to detect individuals in an asymptomatic screening program and can help improve voluntary screening compliance for those individuals averse to various forms of nasal collections.
Keywords: COVID-19; LAMP; RT-qPCR; SARS-CoV-2; oral cavity swab; passive drool; pooling; saliva.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Evaluation of self-collected nasal, urine, and saliva samples for molecular detection of SARS-CoV-2 using an EUA approved RT-PCR assay and a laboratory developed LAMP SARS-CoV-2 test.Immun Inflamm Dis. 2024 Jun;12(6):e1285. doi: 10.1002/iid3.1285. Immun Inflamm Dis. 2024. PMID: 38888444 Free PMC article.
-
SARS-CoV-2 detection by fluorescence loop-mediated isothermal amplification with and without RNA extraction.J Infect Chemother. 2021 Feb;27(2):410-412. doi: 10.1016/j.jiac.2020.10.029. Epub 2020 Oct 31. J Infect Chemother. 2021. PMID: 33214073 Free PMC article.
-
Highly sensitive extraction-free saliva-based molecular assay for rapid diagnosis of SARS-CoV-2.J Clin Microbiol. 2024 Jun 12;62(6):e0060024. doi: 10.1128/jcm.00600-24. Epub 2024 May 24. J Clin Microbiol. 2024. PMID: 38785448 Free PMC article.
-
Screening for SARS-CoV-2 by RT-PCR: Saliva or nasopharyngeal swab? Rapid review and meta-analysis.PLoS One. 2021 Jun 10;16(6):e0253007. doi: 10.1371/journal.pone.0253007. eCollection 2021. PLoS One. 2021. PMID: 34111196 Free PMC article.
-
Evaluation of saliva as a complementary technique to the diagnosis of COVID-19: a systematic review.Med Oral Patol Oral Cir Bucal. 2021 Jul 1;26(4):e526-e532. doi: 10.4317/medoral.24424. Med Oral Patol Oral Cir Bucal. 2021. PMID: 33609022 Free PMC article.
Cited by
-
Development and implementation of a simple and rapid extraction-free saliva SARS-CoV-2 RT-LAMP workflow for workplace surveillance.PLoS One. 2022 May 26;17(5):e0268692. doi: 10.1371/journal.pone.0268692. eCollection 2022. PLoS One. 2022. PMID: 35617204 Free PMC article.
-
Recent advances in RNA sample preparation techniques for the detection of SARS-CoV-2 in saliva and gargle.Trends Analyt Chem. 2023 Aug;165:117107. doi: 10.1016/j.trac.2023.117107. Epub 2023 May 23. Trends Analyt Chem. 2023. PMID: 37317683 Free PMC article. Review.
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
-
Strategies That Facilitate Extraction-Free SARS-CoV-2 Nucleic Acid Amplification Tests.Viruses. 2022 Jun 15;14(6):1311. doi: 10.3390/v14061311. Viruses. 2022. PMID: 35746782 Free PMC article. Review.
-
Development and Validation of Reverse Transcriptase Loop-Mediated Isothermal Amplification (RT-LAMP) as a Simple and Rapid Diagnostic Tool for SARS-CoV-2 Detection.Diagnostics (Basel). 2022 Sep 15;12(9):2232. doi: 10.3390/diagnostics12092232. Diagnostics (Basel). 2022. PMID: 36140632 Free PMC article.
References
-
- Centers for Disease Control and Prevention (CDC) Interim Guidelines for Collecting and Handling of Clinical Specimens for COVID-19 Testing. [(accessed on 10 September 2020)]; Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specim....
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

