A New Class of Uracil-DNA Glycosylase Inhibitors Active against Human and Vaccinia Virus Enzyme
- PMID: 34771075
- PMCID: PMC8587785
- DOI: 10.3390/molecules26216668
A New Class of Uracil-DNA Glycosylase Inhibitors Active against Human and Vaccinia Virus Enzyme
Abstract
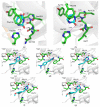
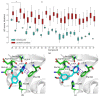
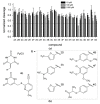
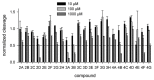
Uracil-DNA glycosylases are enzymes that excise uracil bases appearing in DNA as a result of cytosine deamination or accidental dUMP incorporation from the dUTP pool. The activity of Family 1 uracil-DNA glycosylase (UNG) activity limits the efficiency of antimetabolite drugs and is essential for virulence in some bacterial and viral infections. Thus, UNG is regarded as a promising target for antitumor, antiviral, antibacterial, and antiprotozoal drugs. Most UNG inhibitors presently developed are based on the uracil base linked to various substituents, yet new pharmacophores are wanted to target a wide range of UNGs. We have conducted virtual screening of a 1,027,767-ligand library and biochemically screened the best hits for the inhibitory activity against human and vaccinia virus UNG enzymes. Although even the best inhibitors had IC50 ≥ 100 μM, they were highly enriched in a common fragment, tetrahydro-2,4,6-trioxopyrimidinylidene (PyO3). In silico, PyO3 preferably docked into the enzyme's active site, and in kinetic experiments, the inhibition was better consistent with the competitive mechanism. The toxicity of two best inhibitors for human cells was independent of the presence of methotrexate, which is consistent with the hypothesis that dUMP in genomic DNA is less toxic for the cell than strand breaks arising from the massive removal of uracil. We conclude that PyO3 may be a novel pharmacophore with the potential for development into UNG-targeting agents.
Keywords: DNA repair; inhibitors; pyrimidines; uracil–DNA glycosylase; virtual screening.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Friedberg E.C., Walker G.C., Siede W., Wood R.D., Schultz R.A., Ellenberger T. DNA Repair and Mutagenesis. ASM Press; Washington, DC, USA: 2006. p. 1118.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

