Molecular Classification of Breast Cancer Utilizing Long Non-Coding RNA (lncRNA) Transcriptomes Identifies Novel Diagnostic lncRNA Panel for Triple-Negative Breast Cancer
- PMID: 34771513
- PMCID: PMC8582428
- DOI: 10.3390/cancers13215350
Molecular Classification of Breast Cancer Utilizing Long Non-Coding RNA (lncRNA) Transcriptomes Identifies Novel Diagnostic lncRNA Panel for Triple-Negative Breast Cancer
Abstract
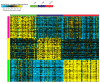
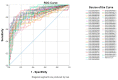
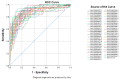
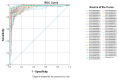
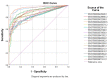
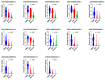
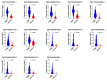
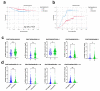
Breast cancer remains the world's most prevalent cancer, responsible for around 685,000 deaths globally despite international research efforts and advances in clinical management. While estrogen receptor positive (ER+), progesterone receptor positive (PR+), and human epidermal growth factor receptor positive (HER2+) subtypes are easily classified and can be targeted, there remains no direct diagnostic test for triple-negative breast cancer (TNBC), except for the lack of receptors expression. The identification of long non-coding RNAs (lncRNAs) and the roles they play in cancer progression has recently proven to be beneficial. In the current study, we utilize RNA sequencing data to identify lncRNA-based biomarkers associated with TNBC, ER+ subtypes, and normal breast tissue. The Marker Finder algorithm identified the lncRNA transcript panel most associated with each molecular subtype and the receiver operating characteristic (ROC) analysis was used to validate the diagnostic potential (area under the curve (AUC) of ≥8.0 and p value < 0.0001). Focusing on TNBC, findings from the discovery cohort were validated in an additional two cohorts, identifying 13 common lncRNA transcripts enriched in TNBC. Binary regression analysis identified a four lncRNA transcript signature (ENST00000425820.1, ENST00000448208.5, ENST00000521666.1, and ENST00000650510.1) with the highest diagnostic power for TNBC. The ENST00000671612.1 lncRNA transcript correlated with worse refractory free survival (RFS). Our data provides a step towards finding a novel diagnostic lncRNA-based panel for TNBC with potential therapeutic implications.
Keywords: TNBC; diagnosis; gene signature; lncRNA; long non-coding RNA; triple-negative breast cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- World Health Organisation Breast Cancer. (Online) Who.int. 2021. [(accessed on 25 October 2021)]. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer.
-
- Grizzi G., Ghidini M., Botticelli A., Tomasello G., Ghidini A., Grossi F., Fusco N., Cabiddu M., Savio T., Petrelli F. Strategies for Increasing the Effectiveness of Aromatase Inhibitors in Locally Advanced Breast Cancer: An Evidence-Based Review on Current Options. Cancer Manag. Res. 2020;12:675–686. doi: 10.2147/CMAR.S202965. - DOI - PMC - PubMed
-
- Llombart-Cussac A., Cortes J., Pare L., Galvan P., Bermejo B., Martinez N., Vidal M., Pernas S., Lopez R., Munoz M., et al. HER2-enriched subtype as a predictor of pathological complete response following trastuzumab and lapatinib without chemotherapy in early-stage HER2-positive breast cancer (PAMELA): An open-label, single-group, multicentre, phase 2 trial. Lancet Oncol. 2017;18:545–554. doi: 10.1016/S1470-2045(17)30021-9. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

