Individualized Proteogenomics Reveals the Mutational Landscape of Melanoma Patients in Response to Immunotherapy
- PMID: 34771574
- PMCID: PMC8582461
- DOI: 10.3390/cancers13215411
Individualized Proteogenomics Reveals the Mutational Landscape of Melanoma Patients in Response to Immunotherapy
Abstract
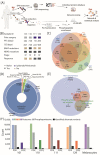
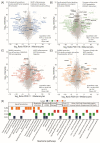
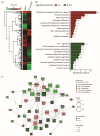
Immune checkpoint inhibitors are used to restore or augment antitumor immune responses and show great promise in the treatment of melanoma and other types of cancers. However, only a small percentage of patients are fully responsive to immune checkpoint inhibition, mostly due to tumor heterogeneity and primary resistance to therapy. Both of these features are largely driven by the accumulation of patient-specific mutations, pointing to the need for personalized approaches in diagnostics and immunotherapy. Proteogenomics integrates patient-specific genomic and proteomic data to study cancer development, tumor heterogeneity and resistance mechanisms. Using this approach, we characterized the mutational landscape of four clinical melanoma patients. This enabled the quantification of hundreds of sample-specific amino acid variants, among them many that were previously not reported in melanoma. Changes in abundance at the protein and phosphorylation site levels revealed patient-specific over-represented pathways, notably linked to melanoma development (MAPK1 activation) or immunotherapy (NLRP1 inflammasome). Personalized data integration resulted in the prediction of protein drug targets, such as the drugs vandetanib and bosutinib, which were experimentally validated and led to a reduction in the viability of tumor cells. Our study emphasizes the potential of proteogenomic approaches to study personalized mutational landscapes, signaling networks and therapy options.
Keywords: immunotherapy; mass spectrometry; melanoma; proteogenomics; whole exome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Proteogenomic Landscape of Breast Cancer Tumorigenesis and Targeted Therapy.Cell. 2020 Nov 25;183(5):1436-1456.e31. doi: 10.1016/j.cell.2020.10.036. Epub 2020 Nov 18. Cell. 2020. PMID: 33212010 Free PMC article.
-
A proteogenomic approach to target neoantigens in solid tumors.Expert Rev Proteomics. 2020 Nov-Dec;17(11-12):797-812. doi: 10.1080/14789450.2020.1881889. Expert Rev Proteomics. 2020. PMID: 33491499 Review.
-
Pan-cancer proteogenomics characterization of tumor immunity.Cell. 2024 Feb 29;187(5):1255-1277.e27. doi: 10.1016/j.cell.2024.01.027. Epub 2024 Feb 14. Cell. 2024. PMID: 38359819 Free PMC article.
-
Proteogenomic Profiling of Treatment-Naïve Metastatic Malignant Melanoma.Cancers (Basel). 2025 Feb 27;17(5):832. doi: 10.3390/cancers17050832. Cancers (Basel). 2025. PMID: 40075679 Free PMC article.
-
Biological Factors behind Melanoma Response to Immune Checkpoint Inhibitors.Int J Mol Sci. 2020 Jun 6;21(11):4071. doi: 10.3390/ijms21114071. Int J Mol Sci. 2020. PMID: 32517213 Free PMC article. Review.
Cited by
-
Deep learning-based multimodal spatial transcriptomics analysis for cancer.Adv Cancer Res. 2024;163:1-38. doi: 10.1016/bs.acr.2024.08.001. Epub 2024 Aug 22. Adv Cancer Res. 2024. PMID: 39271260 Free PMC article. Review.
-
Advances in mechanisms and challenges in clinical translation of synergistic nanomaterial-based therapies for melanoma.Front Cell Dev Biol. 2025 Jul 25;13:1648379. doi: 10.3389/fcell.2025.1648379. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40787624 Free PMC article. Review.
References
-
- Rajasagi M., Shukla S.A., Fritsch E.F., Keskin D.B., DeLuca D., Carmona E., Zhang W., Sougnez C., Cibulskis K., Sidney J., et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014;124:453–462. doi: 10.1182/blood-2014-04-567933. - DOI - PMC - PubMed
-
- Allen E.M.V., Wagle N., Sucker A., Treacy D.J., Johannessen C.M., Goetz E.M., Place C.S., Taylor-Weiner A., Whittaker S., Kryukov G.V., et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melnaoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

