Clinical Value of NGS Genomic Studies for Clinical Management of Pediatric and Young Adult Bone Sarcomas
- PMID: 34771600
- PMCID: PMC8582364
- DOI: 10.3390/cancers13215436
Clinical Value of NGS Genomic Studies for Clinical Management of Pediatric and Young Adult Bone Sarcomas
Abstract
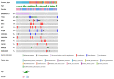
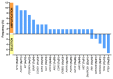
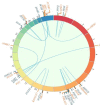
Genomic techniques enable diagnosis and management of children and young adults with sarcomas by identifying high-risk patients and those who may benefit from targeted therapy or participation in clinical trials. Objective: to analyze the performance of an NGS gene panel for the clinical management of pediatric sarcoma patients. We studied 53 pediatric and young adult patients diagnosed with sarcoma, from two Spanish centers. Genomic data were obtained using the Oncomine Childhood Cancer Research Assay, and categorized according to their diagnostic, predictive, or prognostic value. In 44 (83%) of the 53 patients, at least one genetic alteration was identified. In 80% of these patients, the diagnosis was obtained (n = 11) or changed (n = 9), and thus genomic data affected therapy. The most frequent initial misdiagnosis was Ewing's sarcoma, instead of myxoid liposarcoma (FUS-DDDIT3), rhabdoid soft tissue tumor (SMARCB1), or angiomatoid fibrous histiocytoma (EWSR1-CREB1). In our series, two patients had a genetic alteration with an FDA-approved targeted therapy, and 30% had at least one potentially actionable alteration. NGS-based genomic studies are useful and feasible in diagnosis and clinical management of pediatric sarcomas. Genomic characterization of these rare and heterogeneous tumors also helps in the search for prognostic biomarkers and therapeutic opportunities.
Keywords: NGS; genomics; pediatric sarcomas; personalized medicine; targeted therapy.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
References
-
- Peris-Bonet R., Salmerón D., Martínez-Beneito M.A., Galceran J., Marcos-Gragera R., Felipe S., González V., Sánchez de Toledo Codina J. Spanish Childhood Cancer Epidemiology Working Group. Spanish Childhood Cancer Epidemiology Working Group. Childhood cancer incidence and survival in Spain. Ann. Oncol. 2010;21:iii103–iii110. doi: 10.1093/annonc/mdq092. - DOI - PubMed
-
- Gatta G., Botta L., Rossi S., Aareleid T., Bielska-Lasota M., Clavel J., Dimitrova N., Jakab Z., Kaatsch P., Lacour B., et al. Childhood cancer survival in Europe 1999–2007: Results of EUROCARE-5—A population-based study. Lancet Oncol. 2014;15:35–47. doi: 10.1016/S1470-2045(13)70548-5. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

