The Modes of Dysregulation of the Proto-Oncogene T-Cell Leukemia/Lymphoma 1A
- PMID: 34771618
- PMCID: PMC8582492
- DOI: 10.3390/cancers13215455
The Modes of Dysregulation of the Proto-Oncogene T-Cell Leukemia/Lymphoma 1A
Abstract
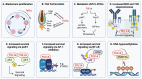
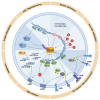
Incomplete biological concepts in lymphoid neoplasms still dictate to a large extent the limited availability of efficient targeted treatments, which entertains the mostly unsatisfactory clinical outcomes. Aberrant expression of the embryonal and lymphatic TCL1 family of oncogenes, i.e., the paradigmatic TCL1A, but also TML1 or MTCP1, is causally implicated in T- and B-lymphocyte transformation. TCL1A also carries prognostic information in these particular T-cell and B-cell tumors. More recently, the TCL1A oncogene has been observed also in epithelial tumors as part of oncofetal stemness signatures. Although the concepts on the modes of TCL1A dysregulation in lymphatic neoplasms and solid tumors are still incomplete, there are recent advances in defining the mechanisms of its (de)regulation. This review presents a comprehensive overview of TCL1A expression in tumors and the current understanding of its (dys)regulation via genomic aberrations, epigenetic modifications, or deregulation of TCL1A-targeting micro RNAs. We also summarize triggers that act through such transcriptional and translational regulation, i.e., altered signals by the tumor microenvironment. A refined mechanistic understanding of these modes of dysregulations together with improved concepts of TCL1A-associated malignant transformation can benefit future approaches to specifically interfere in TCL1A-initiated or -driven tumorigenesis.
Keywords: BPDCN; CLL; T-PLL; TCL1 oncogenes; kinase signaling; lymphoma.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

