Defining genome-wide CRISPR-Cas genome-editing nuclease activity with GUIDE-seq
- PMID: 34773119
- PMCID: PMC9331158
- DOI: 10.1038/s41596-021-00626-x
Defining genome-wide CRISPR-Cas genome-editing nuclease activity with GUIDE-seq
Abstract
Genome-wide unbiased identification of double-stranded breaks enabled by sequencing (GUIDE-seq) is a sensitive, unbiased, genome-wide method for defining the activity of genome-editing nucleases in living cells. GUIDE-seq is based on the principle of efficient integration of an end-protected double-stranded oligodeoxynucleotide tag into sites of nuclease-induced DNA double-stranded breaks, followed by amplification of tag-containing genomic DNA molecules and high-throughput sequencing. Here we describe a detailed GUIDE-seq protocol including cell transfection, library preparation, sequencing and bioinformatic analysis. The entire protocol including cell culture can be completed in 9 d. Once tag-integrated genomic DNA is isolated, library preparation, sequencing and analysis can be performed in 3 d. The result is a genome-wide catalog of off-target sites ranked by nuclease activity as measured by GUIDE-seq read counts. GUIDE-seq is one of the most sensitive cell-based methods for defining genome-wide off-target activity and has been broadly adopted for research and therapeutic use.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Figures
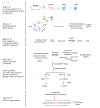

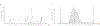

Similar articles
-
Defining CRISPR-Cas9 genome-wide nuclease activities with CIRCLE-seq.Nat Protoc. 2018 Nov;13(11):2615-2642. doi: 10.1038/s41596-018-0055-0. Nat Protoc. 2018. PMID: 30341435 Free PMC article.
-
Identifying genome-wide off-target sites of CRISPR RNA-guided nucleases and deaminases with Digenome-seq.Nat Protoc. 2021 Feb;16(2):1170-1192. doi: 10.1038/s41596-020-00453-6. Epub 2021 Jan 18. Nat Protoc. 2021. PMID: 33462439
-
Tag-seq: a convenient and scalable method for genome-wide specificity assessment of CRISPR/Cas nucleases.Commun Biol. 2021 Jul 2;4(1):830. doi: 10.1038/s42003-021-02351-3. Commun Biol. 2021. PMID: 34215845 Free PMC article.
-
Shooting the messenger: RNA-targetting CRISPR-Cas systems.Biosci Rep. 2018 Jun 21;38(3):BSR20170788. doi: 10.1042/BSR20170788. Print 2018 Jun 29. Biosci Rep. 2018. PMID: 29748239 Free PMC article. Review.
-
Evaluating and Enhancing Target Specificity of Gene-Editing Nucleases and Deaminases.Annu Rev Biochem. 2019 Jun 20;88:191-220. doi: 10.1146/annurev-biochem-013118-111730. Epub 2019 Mar 18. Annu Rev Biochem. 2019. PMID: 30883196 Review.
Cited by
-
An engineered hypercompact CRISPR-Cas12f system with boosted gene-editing activity.Nat Chem Biol. 2023 Nov;19(11):1384-1393. doi: 10.1038/s41589-023-01380-9. Epub 2023 Jul 3. Nat Chem Biol. 2023. PMID: 37400536 Free PMC article.
-
Harnessing noncanonical crRNA for highly efficient genome editing.Nat Commun. 2024 May 7;15(1):3823. doi: 10.1038/s41467-024-48012-x. Nat Commun. 2024. PMID: 38714643 Free PMC article.
-
Versatile and efficient genome editing with Neisseria cinerea Cas9.Commun Biol. 2022 Nov 26;5(1):1296. doi: 10.1038/s42003-022-04258-z. Commun Biol. 2022. PMID: 36435853 Free PMC article.
-
A cleavage rule for selection of increased-fidelity SpCas9 variants with high efficiency and no detectable off-targets.Nat Commun. 2023 Sep 16;14(1):5746. doi: 10.1038/s41467-023-41393-5. Nat Commun. 2023. PMID: 37717069 Free PMC article.
-
Engineered CRISPR-Base Editors as a Permanent Treatment for Familial Dysautonomia.bioRxiv [Preprint]. 2024 Dec 20:2024.11.27.625322. doi: 10.1101/2024.11.27.625322. bioRxiv. 2024. PMID: 39651221 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

