DHX9 contributes to the malignant phenotypes of colorectal cancer via activating NF-κB signaling pathway
- PMID: 34773477
- PMCID: PMC11072136
- DOI: 10.1007/s00018-021-04013-3
DHX9 contributes to the malignant phenotypes of colorectal cancer via activating NF-κB signaling pathway
Abstract
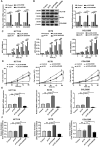
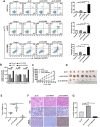
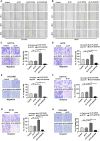
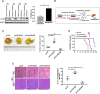
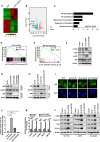
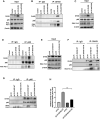
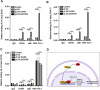
Colorectal cancer (CRC) is the leading cause of cancer-related mortality worldwide, which makes it urgent to identify novel therapeutic targets for CRC treatment. In this study, DHX9 was filtered out as the prominent proliferation promoters of CRC by siRNA screening. Moreover, DHX9 was overexpressed in CRC cell lines, clinical CRC tissues and colitis-associated colorectal cancer (CAC) mouse model. The upregulation of DHX9 was positively correlated with poor prognosis in patients with CRC. Through gain- and loss-of function experiments, we found that DHX9 promoted CRC cell proliferation, colony formation, apoptosis resistance, migration and invasion in vitro. Furthermore, a xenograft mouse model and a hepatic metastasis mouse model were utilized to confirm that forced overexpression of DHX9 enhanced CRC outgrowth and metastasis in vivo, while DHX9 ablation produced the opposite effect. Mechanistically, from one aspect, DHX9 enhances p65 phosphorylation, promotes p65 nuclear translocation to facilitate NF-κB-mediated transcriptional activity. From another aspect, DHX9 interacts with p65 and RNA polymerase II (RNA Pol II) to enhance the downstream targets of NF-κB (e.g., Survivin, Snail) expression to potentiate the malignant phenotypes of CRC. Together, our results suggest that DHX9 may be a potential therapeutic target for prevention and treatment of CRC patients.
Keywords: Colorectal cancer; DHX9; Metastasis; NF-κB signaling; Outgrowth.
© 2021. The Author(s), under exclusive licence to Springer Nature Switzerland AG.
Conflict of interest statement
All authors have no conflict of interest in this paper.
Figures

References
-
- F. Bray, J. Ferlay, I. Soerjomataram, R.L. Siegel, L.A. Torre, A. Jemal, Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA: a cancer journal for clinicians, 68 (2018) 394–424. - PubMed
MeSH terms
Substances
Grants and funding
- 31560260 and 31960163/National Natural Science Foundation of China
- 82003801/National Natural Science Foundation of China
- 20171ACB20024/Natural Science Foundation of Jiangxi Province
- 20181BAB205032/Youth Science Foundation of Jiangxi Province
- TD201703/Technological Innovation Team Project of Gannan Medical University
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

