Comprehensive analyses of bioinformatics applications in the fight against COVID-19 pandemic
- PMID: 34773807
- PMCID: PMC8560182
- DOI: 10.1016/j.compbiolchem.2021.107599
Comprehensive analyses of bioinformatics applications in the fight against COVID-19 pandemic
Abstract
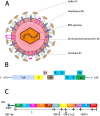
Novel coronavirus disease 2019 (COVID-19) is a global pandemic caused by severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2), which can be transmitted from person to person. As of September 21, 2021, over 228 million cases were diagnosed as COVID-19 infection in more than 200 countries and regions worldwide. The death toll is more than 4.69 million and the mortality rate has reached about 2.05% as it has gradually become a global plague, and the numbers are growing. Therefore, it is important to gain a deeper understanding of the genome and protein characteristics, clinical diagnostics, pathogenic mechanisms, and the development of antiviral drugs and vaccines against the novel coronavirus to deal with the COVID-19 pandemic. The traditional biology technologies are limited for COVID-19-related studies to understand the pandemic happening. Bioinformatics is the application of computational methods and analytical tools in the field of biological research which has obvious advantages in predicting the structure, product, function, and evolution of unknown genes and proteins, and in screening drugs and vaccines from a large amount of sequence information. Here, we comprehensively summarized several of the most important methods and applications relating to COVID-19 based on currently available reports of bioinformatics technologies, focusing on future research for overcoming the virus pandemic. Based on the next-generation sequencing (NGS) and third-generation sequencing (TGS) technology, not only virus can be detected, but also high quality SARS-CoV-2 genome could be obtained quickly. The emergence of data of genome sequences, variants, haplotypes of SARS-CoV-2 help us to understand genome and protein structure, variant calling, mutation, and other biological characteristics. After sequencing alignment and phylogenetic analysis, the bat may be the natural host of the novel coronavirus. Single-cell RNA sequencing provide abundant resource for discovering the mechanism of immune response induced by COVID-19. As an entry receptor, angiotensin-converting enzyme 2 (ACE2) can be used as a potential drug target to treat COVID-19. Molecular dynamics simulation, molecular docking and artificial intelligence (AI) technology of bioinformatics methods based on drug databases for SARS-CoV-2 can accelerate the development of drugs. Meanwhile, computational approaches are helpful to identify suitable vaccines to prevent COVID-19 infection through reverse vaccinology, Immunoinformatics and structural vaccinology.
Keywords: Application; Bioinformatics technology; COVID-19; SARS-CoV-2.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


Similar articles
-
Bioinformatics resources facilitate understanding and harnessing clinical research of SARS-CoV-2.Brief Bioinform. 2021 Mar 22;22(2):714-725. doi: 10.1093/bib/bbaa416. Brief Bioinform. 2021. PMID: 33432321 Free PMC article. Review.
-
A survey on computational methods in discovering protein inhibitors of SARS-CoV-2.Brief Bioinform. 2022 Jan 17;23(1):bbab416. doi: 10.1093/bib/bbab416. Brief Bioinform. 2022. PMID: 34623382 Free PMC article. Review.
-
Molecular characterization, pathogen-host interaction pathway and in silico approaches for vaccine design against COVID-19.J Chem Neuroanat. 2020 Dec;110:101874. doi: 10.1016/j.jchemneu.2020.101874. Epub 2020 Oct 19. J Chem Neuroanat. 2020. PMID: 33091590 Free PMC article. Review.
-
An unprecedented global challenge, emerging trends and innovations in the fight against COVID-19: A comprehensive review.Int J Biol Macromol. 2024 May;267(Pt 1):131324. doi: 10.1016/j.ijbiomac.2024.131324. Epub 2024 Apr 3. Int J Biol Macromol. 2024. PMID: 38574936 Review.
-
Bioinformatics resources for SARS-CoV-2 discovery and surveillance.Brief Bioinform. 2021 Mar 22;22(2):631-641. doi: 10.1093/bib/bbaa386. Brief Bioinform. 2021. PMID: 33416890 Free PMC article.
Cited by
-
Network-Based Data Analysis Reveals Ion Channel-Related Gene Features in COVID-19: A Bioinformatic Approach.Biochem Genet. 2023 Apr;61(2):471-505. doi: 10.1007/s10528-022-10280-x. Epub 2022 Sep 14. Biochem Genet. 2023. PMID: 36104591 Free PMC article.
-
Modern drug discovery applications for the identification of novel candidates for COVID-19 infections.Ann Med Surg (Lond). 2022 Aug;80:104125. doi: 10.1016/j.amsu.2022.104125. Epub 2022 Jul 12. Ann Med Surg (Lond). 2022. PMID: 35845863 Free PMC article. Review.
-
High throughput SARS-CoV-2 variant analysis using molecular barcodes coupled with next generation sequencing.PLoS One. 2022 Jun 21;17(6):e0253404. doi: 10.1371/journal.pone.0253404. eCollection 2022. PLoS One. 2022. PMID: 35727806 Free PMC article.
-
Bioinformatics in Russia: history and present-day landscape.Brief Bioinform. 2024 Sep 23;25(6):bbae513. doi: 10.1093/bib/bbae513. Brief Bioinform. 2024. PMID: 39402695 Free PMC article. Review.
-
Bioinformatics and molecular biology tools for diagnosis, prevention, treatment and prognosis of COVID-19.Heliyon. 2024 Jul 11;10(14):e34393. doi: 10.1016/j.heliyon.2024.e34393. eCollection 2024 Jul 30. Heliyon. 2024. PMID: 39816364 Free PMC article. Review.
References
-
- Beck B.R., Shin B., Choi Y., Park S., Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. - DOI - PMC - PubMed
-
- Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

