Whole-transcriptome sequencing reveals a vernalization-related ceRNA regulatory network in chinese cabbage (Brassica campestris L. ssp. pekinensis)
- PMID: 34773977
- PMCID: PMC8590779
- DOI: 10.1186/s12864-021-08110-2
Whole-transcriptome sequencing reveals a vernalization-related ceRNA regulatory network in chinese cabbage (Brassica campestris L. ssp. pekinensis)
Abstract
Background: The transition from vegetative growth to reproductive growth involves various pathways. Vernalization is a crucial process for floral organ formation and regulation of flowering time that is widely utilized in plant breeding. In this study, we aimed to identify the global landscape of mRNAs, microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) related to vernalization in Chinese cabbage. These data were then used to construct a competitive endogenous RNA (ceRNA) network that provides valuable information to better understand the vernalization response.
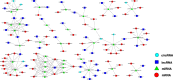
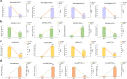
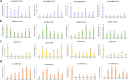
Results: In this study, seeds sampled from the Chinese cabbage doubled haploid (DH) line 'FT' with or without vernalization treatment were used for whole-transcriptome sequencing. A total of 2702 differentially expressed (DE) mRNAs, 151 DE lncRNAs, 16 DE circRNAs, and 233 DE miRNAs were identified in the vernalization-treated seeds. Various transcription factors, such as WRKY, MYB, NAC, bHLH, MADS-box, zinc finger protein CONSTANS-like gene, and B3 domain protein, and regulatory proteins that play important roles in the vernalization pathway were identified. Additionally, we constructed a vernalization-related ceRNA-miRNA-target gene network and obtained 199 pairs of ceRNA relationships, including 108 DEmiRNA‒DEmRNA, 67 DEmiRNA‒DElncRNA, and 12 DEmiRNA‒DEcircRNA interactions, in Chinese cabbage. Furthermore, several important vernalization-related genes and their interacting lncRNAs, circRNAs, and miRNAs, which are involved in the regulation of flowering time, floral organ formation, bolting, and flowering, were identified.
Conclusions: Our results reveal the potential mRNA and non-coding RNAs involved in vernalization, providing a foundation for further studies on the molecular mechanisms underlying vernalization in Chinese cabbage.
Keywords: Chinese cabbage; Non-coding RNA; Vernalization; Whole transcriptome; ceRNA.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







Similar articles
-
Whole transcriptome analysis and construction of a ceRNA regulatory network related to leaf and petiole development in Chinese cabbage (Brassica campestris L. ssp. pekinensis).BMC Genomics. 2023 Mar 24;24(1):144. doi: 10.1186/s12864-023-09239-y. BMC Genomics. 2023. PMID: 36964498 Free PMC article.
-
Whole-transcriptome analysis and construction of an anther development-related ceRNA network in Chinese cabbage (Brassica campestris L. ssp. pekinensis).Sci Rep. 2022 Feb 17;12(1):2667. doi: 10.1038/s41598-022-06556-2. Sci Rep. 2022. PMID: 35177672 Free PMC article.
-
Gene co-expression network analysis reveals key pathways and hub genes in Chinese cabbage (Brassica rapa L.) during vernalization.BMC Genomics. 2021 Apr 6;22(1):236. doi: 10.1186/s12864-021-07510-8. BMC Genomics. 2021. PMID: 33823810 Free PMC article. Review.
-
CircRNA Expression Pattern and ceRNA and miRNA-mRNA Networks Involved in Anther Development in the CMS Line of Brassica campestris.Int J Mol Sci. 2019 Sep 27;20(19):4808. doi: 10.3390/ijms20194808. Int J Mol Sci. 2019. PMID: 31569708 Free PMC article.
-
[Epigenetics of plant vernalization regulated by non-coding RNAs].Yi Chuan. 2012 Jul;34(7):829-34. doi: 10.3724/sp.j.1005.2012.00829. Yi Chuan. 2012. PMID: 22805208 Review. Chinese.
Cited by
-
Regulatory Mechanism of Proanthocyanidins in Grape Peels Using vvi-miR828a and Its Target Gene VvMYBPA1.Plants (Basel). 2024 Jun 18;13(12):1688. doi: 10.3390/plants13121688. Plants (Basel). 2024. PMID: 38931120 Free PMC article.
-
Identification and Validation of Genes Exhibiting Dynamic Alterations in Response to Bleomycin-Induced Pulmonary Fibrosis.Mol Biotechnol. 2024 Nov;66(11):3323-3335. doi: 10.1007/s12033-023-00943-4. Epub 2023 Nov 4. Mol Biotechnol. 2024. PMID: 37924392
-
Increase Crop Resilience to Heat Stress Using Omic Strategies.Front Plant Sci. 2022 May 17;13:891861. doi: 10.3389/fpls.2022.891861. eCollection 2022. Front Plant Sci. 2022. PMID: 35656008 Free PMC article. Review.
-
The Sink-Source Relationship in Cucumber (Cucumis sativus L.) Is Modulated by DNA Methylation.Plants (Basel). 2023 Dec 28;13(1):103. doi: 10.3390/plants13010103. Plants (Basel). 2023. PMID: 38202411 Free PMC article.
-
Source leaves are regulated by sink strengths through non-coding RNAs and alternative polyadenylation in cucumber (Cucumis sativus L.).BMC Plant Biol. 2024 Aug 29;24(1):812. doi: 10.1186/s12870-024-05416-7. BMC Plant Biol. 2024. PMID: 39198785 Free PMC article.
References
-
- Amasino R. Seasonal and developmental timing of flowering. Plant J. 2010;61(6):1001–13. - PubMed
-
- Putterill J, Laurie R, Macknight R. It’s time to flower: the genetic control of flowering time. BioEssays. 2004;26(4):363–73. - PubMed
-
- Jung C, Müller AE. Flowering time control and applications in plant breeding. Trends Plant Sci. 2009;14(10):563–73. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

