Structural and functional significance of the amino acid differences Val35Thr, Ser46Ala, Asn65Ser, and Ala94Ser in 3C-like proteinases from SARS-CoV-2 and SARS-CoV
- PMID: 34774600
- PMCID: PMC8580570
- DOI: 10.1016/j.ijbiomac.2021.11.043
Structural and functional significance of the amino acid differences Val35Thr, Ser46Ala, Asn65Ser, and Ala94Ser in 3C-like proteinases from SARS-CoV-2 and SARS-CoV
Abstract
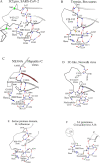
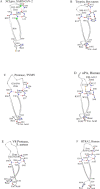
Three dimensional structures of (chymo)trypsin-like proteinase (3CLpro) from SARS-CoV-2 and SARS-CoV differ at 8 positions. We previously found that the Val86Leu, Lys88Arg, Phe134His, and Asn180Lys mutations in these enzymes can change the orientation of the N- and C-terminal domains of 3CLpro relative to each other, which leads to a change in catalytic activity. This conclusion was derived from the comparison of the structural catalytic core in 169 (chymo)trypsin-like proteinases with the serine/cysteine fold. Val35Thr, Ser46Ala, Asn65Ser, Ala94Ser mutations were not included in that analysis, since they are located far from the catalytic tetrad. In the present work, the structural and functional roles of these variable amino acids at positions 35, 46, 65, and 94 in the 3CLpro sequences of SARS-CoV-2 and SARS-CoV have been established using a comparison of the same set of proteinases leading to the identification of new conservative elements. Comparative analysis showed that, in addition to interdomain mobility, which could modulate catalytic activity, the 3CLpro(s) can use for functional regulation an autolytic loop and the unique Asp33-Asn95 region (the Asp33-Asn95 Zone) in the N-terminal domain. Therefore, all 4 analyzed mutation sites are associated with the unique structure-functional features of the 3CLpro from SARS-CoV-2 and SARS-CoV. Strictly speaking, the presented structural results are hypothetical, since at present there is not a single experimental work on the identification and characterization of autolysis sites in these proteases.
Keywords: (Chymo)trypsin-like proteinases; Autolysis; COVID-19; Catalytic tetrad; Interdomain loop; SARS-CoV-2; Structural catalytic core.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Structural leitmotif and functional variations of the structural catalytic core in (chymo)trypsin-like serine/cysteine fold proteinases.Int J Biol Macromol. 2021 May 15;179:601-609. doi: 10.1016/j.ijbiomac.2021.03.042. Epub 2021 Mar 10. Int J Biol Macromol. 2021. PMID: 33713772
-
Allosteric Regulation of 3CL Protease of SARS-CoV-2 and SARS-CoV Observed in the Crystal Structure Ensemble.J Mol Biol. 2021 Dec 3;433(24):167324. doi: 10.1016/j.jmb.2021.167324. Epub 2021 Oct 27. J Mol Biol. 2021. PMID: 34717972 Free PMC article.
-
Enzymatic activity characterization of SARS coronavirus 3C-like protease by fluorescence resonance energy transfer technique.Acta Pharmacol Sin. 2005 Jan;26(1):99-106. doi: 10.1111/j.1745-7254.2005.00010.x. Acta Pharmacol Sin. 2005. PMID: 15659121 Free PMC article.
-
Design and Evaluation of Anti-SARS-Coronavirus Agents Based on Molecular Interactions with the Viral Protease.Molecules. 2020 Aug 27;25(17):3920. doi: 10.3390/molecules25173920. Molecules. 2020. PMID: 32867349 Free PMC article. Review.
-
An Updated Review on SARS-CoV-2 Main Proteinase (MPro): Protein Structure and Small-Molecule Inhibitors.Curr Top Med Chem. 2021;21(6):442-460. doi: 10.2174/1568026620666201207095117. Curr Top Med Chem. 2021. PMID: 33292134 Review.
Cited by
-
The SARS-CoV-2 main protease (Mpro): Structure, function, and emerging therapies for COVID-19.MedComm (2020). 2022 Jul 14;3(3):e151. doi: 10.1002/mco2.151. eCollection 2022 Sep. MedComm (2020). 2022. PMID: 35845352 Free PMC article. Review.
-
Surface cysteines could protect the SARS-CoV-2 main protease from oxidative damage.J Inorg Biochem. 2022 Sep;234:111886. doi: 10.1016/j.jinorgbio.2022.111886. Epub 2022 Jun 2. J Inorg Biochem. 2022. PMID: 35675741 Free PMC article.
References
-
- Hegyi A., Ziebuhr J. Conservation of substrate specificities among coronavirus main proteases. J. Gen. Virol. 2002;83(Pt 3):595–599. - PubMed
-
- Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 2003;84(Pt 9):2305–2315. - PubMed
-
- Yang H., Yang M., Ding Y., Liu Y., Lou Z., Zhou Z., Sun L., Mo L., Ye S., Pang H., Gao G.F., Anand K., Bartlam M., Hilgenfeld R., Rao Z. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. U. S. A. 2003;100(23):13190–13195. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

