SARS-CoV-2 S1 Protein Induces Endolysosome Dysfunction and Neuritic Dystrophy
- PMID: 34776872
- PMCID: PMC8579006
- DOI: 10.3389/fncel.2021.777738
SARS-CoV-2 S1 Protein Induces Endolysosome Dysfunction and Neuritic Dystrophy
Abstract
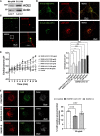
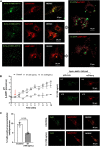
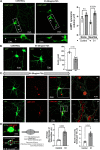
SARS-CoV-2 is the viral cause of the COVID-19 pandemic. Increasingly, significant neurological disorders have been associated with COVID-19. However, the pathogenesis of these neurological disorders remains unclear especially because only low or undetectable levels of SARS-CoV-2 have been reported in human brain specimens. Because SARS-CoV-2 S1 protein can be released from viral membranes, can cross the blood-brain barrier, and is present in brain cells including neurons, we tested the hypothesis that SARS-CoV-2 S1 protein can directly induce neuronal injury. Incubation of primary human cortical neurons with SARS-CoV-2 S1 protein resulted in accumulation of the S1 protein in endolysosomes as well as endolysosome de-acidification. Further, SARS-CoV-2 S1 protein induced aberrant endolysosome morphology and neuritic varicosities. Our findings suggest that SARS-CoV-2 S1 protein directly induces neuritic dystrophy, which could contribute to the high incidence of neurological disorders associated with COVID-19.
Keywords: SARS-CoV-2 S1; endocytosis; endolysosomal acidification; lysosome; neuritic dystrophy; neuron.
Copyright © 2021 Datta, Miller, Halcrow, Khan, Colwell, Geiger and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
SLC38A9 regulates SARS-CoV-2 viral entry.iScience. 2024 Jun 27;27(7):110387. doi: 10.1016/j.isci.2024.110387. eCollection 2024 Jul 19. iScience. 2024. PMID: 39071889 Free PMC article.
-
HIV-1 gp120-Induced Endolysosome de-Acidification Leads to Efflux of Endolysosome Iron, and Increases in Mitochondrial Iron and Reactive Oxygen Species.J Neuroimmune Pharmacol. 2022 Jun;17(1-2):181-194. doi: 10.1007/s11481-021-09995-2. Epub 2021 Apr 8. J Neuroimmune Pharmacol. 2022. PMID: 33834418 Free PMC article.
-
Antibodies against the SARS-CoV-2 S1-RBD cross-react with dengue virus and hinder dengue pathogenesis.Front Immunol. 2022 Aug 15;13:941923. doi: 10.3389/fimmu.2022.941923. eCollection 2022. Front Immunol. 2022. PMID: 36045680 Free PMC article.
-
SARS-CoV-2 Infection of Human Neurons Is TMPRSS2 Independent, Requires Endosomal Cell Entry, and Can Be Blocked by Inhibitors of Host Phosphoinositol-5 Kinase.J Virol. 2023 Apr 27;97(4):e0014423. doi: 10.1128/jvi.00144-23. Epub 2023 Apr 11. J Virol. 2023. PMID: 37039676 Free PMC article.
-
Neurological manifestations of SARS-CoV-2: complexity, mechanism and associated disorders.Eur J Med Res. 2023 Aug 30;28(1):307. doi: 10.1186/s40001-023-01293-2. Eur J Med Res. 2023. PMID: 37649125 Free PMC article. Review.
Cited by
-
The Endolysosomal System: The Acid Test for SARS-CoV-2.Int J Mol Sci. 2022 Apr 20;23(9):4576. doi: 10.3390/ijms23094576. Int J Mol Sci. 2022. PMID: 35562967 Free PMC article. Review.
-
SLC38A9 is directly involved in Tat-induced endolysosome dysfunction and senescence in astrocytes.Life Sci Alliance. 2025 May 5;8(7):e202503231. doi: 10.26508/lsa.202503231. Print 2025 Jul. Life Sci Alliance. 2025. PMID: 40324823 Free PMC article.
-
Molecular Mechanisms in the Genesis of Seizures and Epilepsy Associated With Viral Infection.Front Mol Neurosci. 2022 May 9;15:870868. doi: 10.3389/fnmol.2022.870868. eCollection 2022. Front Mol Neurosci. 2022. PMID: 35615063 Free PMC article. Review.
-
Dimethoxycurcumin Acidifies Endolysosomes and Inhibits SARS-CoV-2 Entry.Front Virol. 2022;2:923018. doi: 10.3389/fviro.2022.923018. Epub 2022 Jun 29. Front Virol. 2022. PMID: 39677976 Free PMC article.
-
TLR7 Mediates HIV-1 Tat-Induced Cellular Senescence in Human Astrocytes.Aging Cell. 2025 Jul;24(7):e70086. doi: 10.1111/acel.70086. Epub 2025 Apr 30. Aging Cell. 2025. PMID: 40304459 Free PMC article.
References
-
- Buzhdygan T. P., Deore B. J., Baldwin-Leclair A., Mcgary H., Razmpour R., Galie P. A., et al. (2020). The SARS-CoV-2 spike protein alters barrier function in 2D static and 3D microfluidic in vitro models of the human blood–brain barrier. Neurobiol. Dis. 146:105131. 10.1016/j.nbd.2020.105131 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

