Biosensor-Based Optimization of Cutinase Secretion by Corynebacterium glutamicum
- PMID: 34777299
- PMCID: PMC8581548
- DOI: 10.3389/fmicb.2021.750150
Biosensor-Based Optimization of Cutinase Secretion by Corynebacterium glutamicum
Abstract
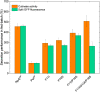
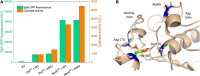
The industrial microbe Corynebacterium glutamicum is gaining substantial importance as a platform host for recombinant protein secretion. We recently developed a fluorescence-based (eYFP) C. glutamicum reporter strain for the quantification of Sec-dependent protein secretion by monitoring the secretion-related stress response and now demonstrate its applicability in optimizing the secretion of the heterologous enzyme cutinase from Fusarium solani pisi. To drive secretion, either the poor-performing PelSP or the potent NprESP Sec signal peptide from Bacillus subtilis was used. To enable easy detection and quantification of the secreted cutinase we implemented the split green fluorescent protein (GFP) assay, which relies on the GFP11-tag fused to the C-terminus of the cutinase, which can complement a truncated GFP thereby reconstituting its fluorescence. The reporter strain was transformed with different mutant libraries created by error-prone PCR, which covered the region of the signal peptide and the N-terminus of the cutinase. Fluorescence-activated cell sorting (FACS) was performed to isolate cells that show increased fluorescence in response to increased protein secretion stress. Five PelSP variants were identified that showed a 4- to 6-fold increase in the amount and activity of the secreted cutinase (up to 4,100 U/L), whereas two improved NprESP variants were identified that showed a ∼35% increase in secretion, achieving ∼5,500 U/L. Most of the isolated variants carried mutations in the h-region of the signal peptide that increased its overall hydrophobicity. Using site-directed mutagenesis it was shown that the combined mutations F11I and P16S within the hydrophobic core of the PelSP are sufficient to boost cutinase secretion in batch cultivations to the same level as achieved by the NprESP. Screening of a PelSP mutant library in addition resulted in the identification of a cutinase variant with an increased specific activity, which was attributed to the mutation A85V located within the substrate-binding region. Taken together the biosensor-based optimization approach resulted in a substantial improvement of cutinase secretion by C. glutamicum, and therefore represents a valuable tool that can be applied to any secretory protein of interest.
Keywords: Corynebacterium glutamicum; EYFP; FACS; Sec-dependent export; fluorescence-based biosensor; protein secretion; signal peptide; split GFP.
Copyright © 2021 Bakkes, Lenz, Müller, Bida, Dohmen-Olma, Knapp, Oldiges, Jaeger and Freudl.
Conflict of interest statement
AK is employed by Castrol Germany GmbH. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Adams H., Scotti P. A., De Cock H., Luirink J., Tommassen J. (2002). The presence of a helix breaker in the hydrophobic core of signal sequences of secretory proteins prevents recognition by the signal-recognition particle in Escherichia coli. Eur. J. Biochem. 269 5564–5571. 10.1046/j.1432-1033.2002.03262.x - DOI - PubMed
-
- Brockmeier U., Caspers M., Freudl R., Jockwer A., Noll T., Eggert T. (2006). Systematic screening of all signal peptides from Bacillus subtilis: a powerful strategy in optimizing heterologous protein secretion in Gram-positive bacteria. J. Mol. Biol. 362 393–402. 10.1016/j.jmb.2006.07.034 - DOI - PubMed
LinkOut - more resources
Full Text Sources

