An OsKala3, R2R3 MYB TF, Is a Common Key Player for Black Rice Pericarp as Main Partner of an OsKala4, bHLH TF
- PMID: 34777449
- PMCID: PMC8585765
- DOI: 10.3389/fpls.2021.765049
An OsKala3, R2R3 MYB TF, Is a Common Key Player for Black Rice Pericarp as Main Partner of an OsKala4, bHLH TF
Abstract
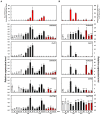
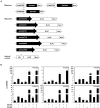
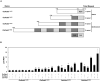
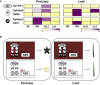
Rice (Oryza sativa) pericarp exhibits various colors due to the accumulation of anthocyanins and/or proanthocyanidins. Previous work revealed that the two basic helix-loop-helix (bHLH) transcription factors OsKala4 and OsRc are key regulators for the black and red pericarp traits, respectively, and their inactivation results in rice with white pericarp. However, their pericarp-specific R2R3 MYB partner remained unknown. Here, we characterized the role of the R2R3 MYB gene OsKala3 in rice pericarp pigmentation through genetic and molecular approaches. A rice protoplast transfection assay showed that OsKala3 is a nuclear-localized protein. Furthermore, OsKala3 physically interacted with OsKala4 in a yeast two-hybrid analysis. Co-transfection assays in rice protoplasts revealed that OsKala3 and OsKala4 mediate the activation of anthocyanin biosynthetic genes. Notably, the OsKala3 promoter region exhibited an insertion polymorphism specifically in rice cultivars with black pericarp, creating two tandem repeats while red and white varieties harbor only one. The number of repeats within the OsKala3 promoter correlated with increased transactivation by OsKala3, thus providing a rationale for the black pericarp characteristic of cultivars with two repeats. These results thus provide evidence for the molecular basis of anthocyanin biosynthesis in rice pericarp and may facilitate the introduction of this beneficial trait to other rice cultivars through marker-assisted breeding.
Keywords: OsKala3; OsKala4; anthocyanin; regulation system; rice; tandem repeats in promoters.
Copyright © 2021 Kim, Yang, Ha, Kim, Lee and Lim.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Fan F. J., Fan Y. Y., Du J. H., Zhuang J. Y. (2008). Fine mapping of C (Chromogen for anthocyanin) gene in rice. Rice Sci. 15 1–6. 10.1016/S1672-6308(08)60012-8 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

