RNA Biological Characteristics at the Peak of Cell Death in Different Hereditary Retinal Degeneration Mutants
- PMID: 34777465
- PMCID: PMC8586524
- DOI: 10.3389/fgene.2021.728791
RNA Biological Characteristics at the Peak of Cell Death in Different Hereditary Retinal Degeneration Mutants
Abstract
Purpose: The present work investigated changes in the gene expression, molecular mechanisms, and pathogenesis of inherited retinal degeneration (RD) in three different disease models, to identify predictive biomarkers for their varied phenotypes and to provide a better scientific basis for their diagnosis, treatment, and prevention. Methods: Differentially expressed genes (DEGs) between retinal tissue from RD mouse models obtained during the photoreceptor cell death peak period (Pde6b rd1 at post-natal (PN) day 13, Pde6b rd10 at PN23, Prph rd2 at PN29) and retinal tissue from C3H wild-type mice were identified using Illumina high-throughput RNA-sequencing. Co-expression gene modules were identified using a combination of GO and KEGG enrichment analyses and gene co-expression network analysis. CircRNA-miRNA-mRNA network interactions were studied by genome-wide circRNA screening. Results: Pde6b rd1 , Pde6b rd10 , and Prph rd2 mice had 1,926, 3,096, and 375 DEGs, respectively. Genes related to ion channels, stress, inflammatory processes, tumor necrosis factor (TNF) production, and microglial cell activation were up-regulated, while genes related to endoplasmic reticulum regulation, metabolism, and homeostasis were down-regulated. Differential expression of transcription factors and non-coding RNAs generally implicated in other human diseases was detected (e.g., glaucoma, diabetic retinopathy, and inherited retinal degeneration). CircRNA-miRNA-mRNA network analysis indicated that these factors may be involved in photoreceptor cell death. Moreover, excessive cGMP accumulation causes photoreceptor cell death, and cGMP-related genes were generally affected by different pathogenic gene mutations. Conclusion: We screened genes and pathways related to photoreceptor cell death. Additionally, up-stream regulatory factors, such as transcription factors and non-coding RNA and their interaction networks were analyzed. Furthermore, RNAs involved in RD were functionally annotated. Overall, this study lays a foundation for future studies on photoreceptor cell death mechanisms.
Keywords: RNA-seq; biogenic analysis; cGMP-related genes; inherited retinal degeneration (IRD); photoreceptor cell death.
Copyright © 2021 Wei, Li, Feng, Hu, Paquet-Durand and Jiao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
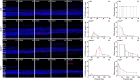


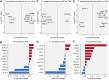
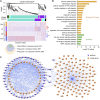
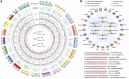

References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

