RNA Sequencing Reveals Phenylpropanoid Biosynthesis Genes and Transcription Factors for Hevea brasiliensis Reaction Wood Formation
- PMID: 34777481
- PMCID: PMC8585928
- DOI: 10.3389/fgene.2021.763841
RNA Sequencing Reveals Phenylpropanoid Biosynthesis Genes and Transcription Factors for Hevea brasiliensis Reaction Wood Formation
Abstract
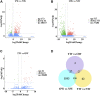
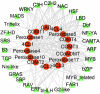
Given the importance of wood in many industrial applications, much research has focused on wood formation, especially lignin biosynthesis. However, the mechanisms governing the regulation of lignin biosynthesis in the rubber tree (Hevea brasiliensis) remain to be elucidated. Here, we gained insight into the mechanisms of rubber tree lignin biosynthesis using reaction wood (wood with abnormal tissue structure induced by gravity or artificial mechanical treatment) as an experimental model. We performed transcriptome analysis of rubber tree mature xylem from tension wood (TW), opposite wood (OW), and normal wood (NW) using RNA sequencing (RNA-seq). A total of 214, 1,280, and 32 differentially expressed genes (DEGs) were identified in TW vs. NW, OW vs. NW, and TW vs. OW, respectively. GO and KEGG enrichment analysis of DEGs from different comparison groups showed that zeatin biosynthesis, plant hormone signal transduction, phenylpropanoid biosynthesis, and plant-pathogen interaction pathways may play important roles in reaction wood formation. Sixteen transcripts involved in phenylpropanoid biosynthesis and 129 transcripts encoding transcription factors (TFs) were used to construct a TF-gene regulatory network for rubber tree lignin biosynthesis. Among them, MYB, C2H2, and NAC TFs could regulate all the DEGs involved in phenylpropanoid biosynthesis. Overall, this study identified candidate genes and TFs likely involved in phenylpropanoid biosynthesis and provides novel insights into the mechanisms regulating rubber tree lignin biosynthesis.
Keywords: lignin biosynthesis; phenylpropanoid biosynthesis pathway; reaction wood; rubber tree; rubber wood.
Copyright © 2021 Meng, Wang, Li, Jiao, Zhang, Zhang, Chen and Tu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Chang S., Puryear J., Cairney J. (1993). A Simple and Efficient Method for Isolating RNA from Pine Trees. Plant Mol. Biol. Rep. 11, 113–116. 10.1007/bf02670468 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

