The Upregulation of PLXDC2 Correlates with Immune Microenvironment Characteristics and Predicts Prognosis in Gastric Cancer
- PMID: 34777633
- PMCID: PMC8589478
- DOI: 10.1155/2021/5669635
The Upregulation of PLXDC2 Correlates with Immune Microenvironment Characteristics and Predicts Prognosis in Gastric Cancer
Abstract
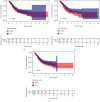
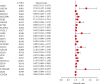
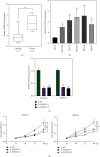
Tumor microenvironment (TME) has been demonstrated to exhibit a regulatory effect on the progressions of gastric cancer (GC). However, the related functions of stromal and immune components (TME-associated genes) in TME remain largely unclear. From the TCGA dataset, we downloaded the clinical data of 375 GC cases and then estimated the percentage of tumor-infiltrating immunocytes (TICs) and the levels of immune and stromal constituents by the use of CIBERSORT and ESTIMATE tolls. Univariate assays were applied to study the differentially expressed genes. The associations between the clinical information of GC patients and the expressions of the specific genes were analyzed based on the TCGA datasets. The effect of Plexin domain containing 2 (PLXDC2) expression on TICs was conducted. We observed that PLXDC2 expression was distinctly upregulated in GC specimens compared with nontumor gastric specimens. Its upregulation was associated with advanced clinical stages and predicted a shorter overall survival of GC patients. The genes in the group of higher expressing PLXDC2 were primarily enriched in immunity-associated events. By the use of CIBERSORT, we observed that PLXDC2 expressions were related to the proportion of dendritic cells resting, T cell CD4 memory resting, eosinophils, mastocyte resting, mononuclear cells, plasma cells, T cell follicle helper, macrophage M2, and dendritic cells activated. Overall, our discoveries revealed that the expression of PLXDC2 was remarkable in GC, might be a possible biomarker for GC, and provided novel contents regarding immune infiltrates, offering novel insight for treatments of GC.
Copyright © 2021 Yang Li et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

